Bacterial Expression
Bacterial expression systems are among the most widely used platforms in molecular biology and biotechnology for the production of recombinant proteins for research, therapeutic and industrial purposes. Their simplicity, cost-effectiveness and versatility make them indispensable tools for scientific progress. Bacterial expression involves the introduction of foreign DNA, typically encoding a protein of interest, into bacterial cells to facilitate the synthesis of that protein. The most commonly used bacterial hosts are Escherichia coli (E. coli) and Bacillus subtilis due to their well-characterized genetics, rapid growth, and ability to achieve high-density cultures.
Learn more about the characteristics of bacterial expression with Creative Biostructure and explore our bacterial expression services.
Bacterial Expression Systems
Several bacterial systems exist, each tailored to specific requirements for protein expression.
Escherichia coli (E. coli)
E. coli is the most widely used bacterial expression host due to its rapid growth, well-understood genetics, and availability of diverse vectors and strains. Expression in E. coli is facilitated by strong promoters such as T7 or lac, ribosome binding sites, and efficient termination sequences. However, challenges such as improper protein folding, aggregation into inclusion bodies, and the inability to perform post-translational modifications (PTMs) remain limitations.
Bacillus subtilis
B. subtilis is another bacterial system used for protein expression. Unlike E. coli, it secretes proteins directly into the culture medium, simplifying downstream processing. It is particularly useful for the production of enzymes and proteins that require extracellular secretion.
Other Bacterial Hosts
Hosts such as Pseudomonas putida and Corynebacterium glutamicum are emerging as alternative systems for specific applications, especially where protein solubility or metabolic compatibility with synthetic pathways is crucial.
Process of Bacterial Expression
The process of bacterial expression involves a series of well-defined steps to produce a target protein within bacterial cells. This process takes advantage of the ability of bacteria to rapidly transcribe and translate genes, allowing for high-yield protein production. Below is an overview of the key steps involved:
Selection of the Gene of Interest
The target gene encoding the desired protein is identified and isolated. The gene sequence is often optimized to match the codon usage preferences of the bacterial host, typically E. coli, for efficient translation. The gene may be chemically synthesized or cloned from natural sources using polymerase chain reaction (PCR) techniques.
Choice of Expression Vector
An appropriate bacterial expression plasmid is selected. These plasmids typically contain: A strong promoter (e.g., T7, lac) to drive gene expression; a ribosome-binding site (RBS) for translation initiation; selectable markers, such as antibiotic resistance genes, to maintain the plasmid in bacterial cultures; and optional fusion tags (e.g., His-tag, GST-tag) for protein purification.
The gene of interest is then inserted into the plasmid using restriction enzyme digestion and ligation or seamless cloning techniques such as Gibson Assembly.
Transformation into Host Cells
The recombinant plasmid is introduced into bacterial host cells, typically E. coli, using one of the following methods:
- Heat Shock Transformation: Cells are made competent with calcium chloride and briefly exposed to heat to allow plasmid uptake.
- Electroporation: An electric pulse creates temporary pores in the bacterial membrane, facilitating plasmid entry.
Screening for Transformants
After transformation, bacteria are plated on selective media containing an antibiotic corresponding to the resistance marker of the plasmid. Only bacteria containing the plasmid grow, allowing selection of successfully transformed colonies.
Optimization of Expression Conditions
The gene of interest is under the control of an inducible promoter, such as the IPTG-inducible T7 promoter. Inducers such as IPTG (isopropyl β-D-1-thiogalactopyranoside) are added to the culture to initiate transcription. Temperature, pH, and media composition are optimized to maximize protein yield while minimizing problems such as aggregation or toxicity. Low temperatures (16-25°C) can be used to promote proper protein folding.
Protein Synthesis
Once induced, the bacterial machinery transcribes the gene into mRNA and translates it into the target protein. The synthesized protein may remain in the cytoplasm, periplasm or be secreted into the culture medium, depending on the signal sequences and the host strain used.
Harvesting and Lysis
Bacteria are collected from the culture by centrifugation to concentrate the cells. Cells are broken open to release the target protein using methods such as physical disruption (sonication or high-pressure homogenization) and chemical lysis (detergents or lysozyme treatment).
Protein Purification
The lysate is clarified by removing cell debris by centrifugation or filtration. Purification is performed by techniques such as affinity chromatography (using fusion tags, e.g., his-tag binding to nickel columns), ion exchange chromatography (separating proteins based on charge), and size exclusion chromatography (isolating proteins based on size).
Refolding and Post-Purification Steps
If the protein forms insoluble inclusion bodies, it is solubilized and refolded into its functional conformation through refolding buffers and dialysis. Further quality control steps, such as SDS-PAGE and Western blotting, confirm the identity and purity of the expressed protein.
Application or Storage
The purified protein is either used directly for downstream applications like structural studies, enzyme assays, or therapeutic development or stored under optimal conditions for future use (e.g., frozen at -80°C or lyophilized).
A simplified high-throughput bacterial expression process is shown below:
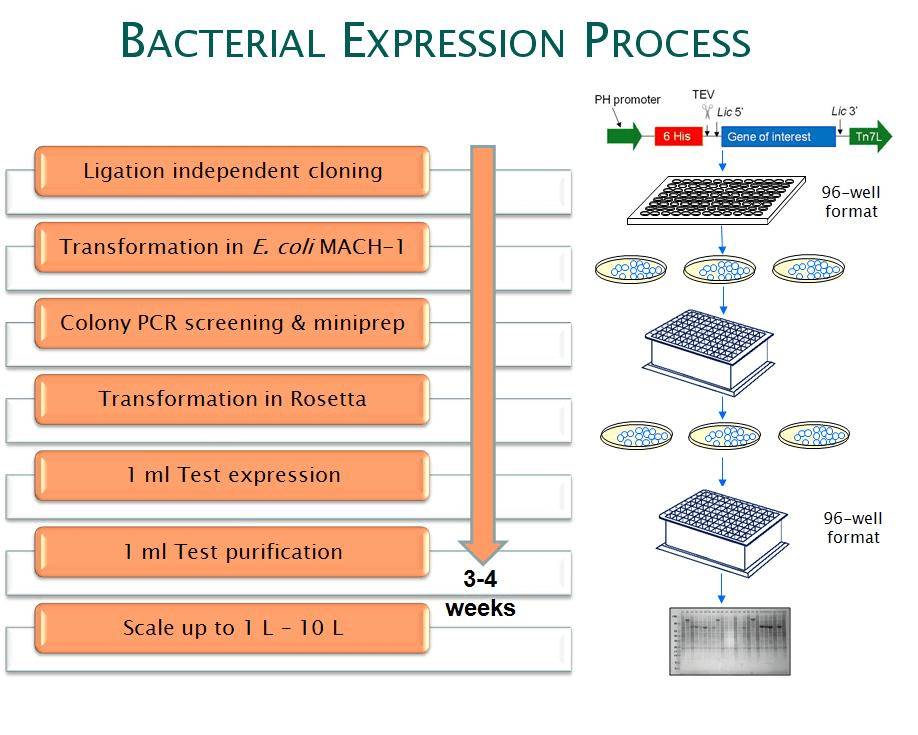 Figure 1. Simplified process of bacterial expression system.
Figure 1. Simplified process of bacterial expression system.
Applications of Bacterial Expression
Recombinant Protein Expression and Purification
Bacterial hosts are essential for both laboratory and large-scale recombinant protein production. By optimizing host strains, expression vectors and downstream processes, high-yield protein production is possible. These proteins are then used for research, diagnostic and therapeutic applications. For example, therapeutic proteins such as insulin and growth hormones are commonly expressed in E. coli.
Heteromultimeric Protein Expression
A heteromultimeric protein is a protein complex that contains multiple different protein subunits. Bacterial systems can express these complex proteins by co-expressing multiple subunits, often using multi-gene expression plasmids. This capability is vital for studying protein-protein interactions and assembling functional protein complexes.
Research Applications
The ability to express recombinant proteins has transformed molecular biology. Bacterial systems allow rapid production of proteins for structural and functional studies, providing insight into enzyme mechanisms, protein-ligand interactions, and cellular pathways. For example, the production of GFP (green fluorescent protein) in E. coli has facilitated cellular imaging and live cell monitoring.
Whole Cell Biocatalysis
Bacterial expression systems are an integral part of whole-cell biocatalysis, where engineered bacteria serve as stable platforms for complex metabolic pathways. These pathways enable the biosynthesis of valuable products such as biofuels, bioplastics, and pharmaceuticals.
Advantages and Limitations of Bacterial Expression
| Advantages of Bacterial Expression Systems | |
| Bacterial Expression | The simple physiology of bacteria simplifies genetic manipulation, transformation and expression processes. Unlike eukaryotic cells, bacteria lack organelles, which ensures that transcription and translation occur in a single compartment, increasing the efficiency of protein synthesis. |
| Short Generation Time | Bacteria multiply rapidly, with doubling times as short as 20 minutes under optimal conditions. This short generation timeline allows researchers to achieve high-density cultures and produce significant amounts of recombinant proteins in a matter of hours to days. |
| High Yields with Controlled Expression | Bacterial systems provide tight regulation of gene expression, often using inducible promoters such as T7, lac or arabinose systems. This allows precise control of protein production and can result in protein yields of up to 10% of the total cell mass. |
| Broad Expression Range | The ability to modulate expression levels using varying concentrations of inducers ensures versatility, making bacterial systems suitable for expressing proteins at low levels to prevent toxicity or at high levels to maximize yield. |
| Temperature and Media Adaptability | Bacterial expression systems can function effectively at low temperatures (as low as 16ºC), which is critical for minimizing protein misfolding and aggregation. They are also less sensitive to variations in media formulations, which reduces costs and streamlines culture optimization. |
| Tag-Based Protein Purification | The availability of vectors with fusion tags, such as His-tags, GST-tags, or MBP-tags, simplifies protein purification. These tags enable the use of low-cost affinity supports for efficient protein isolation and purification. |
| Scalability and Cost Efficiency | Bacterial systems are highly scalable, allowing seamless transition from laboratory-scale experiments to large-scale industrial production. Their minimal nutrient requirements and inexpensive media further enhance their cost-effectiveness. |
| Enhanced Soluble Protein Recovery | Compared to other expression systems, bacterial hosts often achieve higher success rates in producing soluble proteins, especially when optimized expression conditions and solubility-enhancing strategies are employed. |
| Limitations of Bacterial Expression Systems | |
| Improper Protein Folding | Bacteria lack the sophisticated cellular machinery required to properly fold many complex proteins. As a result, expressed proteins can aggregate into insoluble inclusion bodies, rendering them biologically inactive. While it is possible to refold proteins from inclusion bodies, it is often labor-intensive and inefficient. |
| Toxicity to Host Cells | Overexpression of certain proteins can be toxic to bacteria, inhibiting cell growth and preventing cultures from reaching high densities. This challenge can be mitigated by using inducible promoters that delay protein expression until the bacterial culture has grown to a desired density. |
| Lack of Post-Translational Modifications (PTMs) | Bacteria do not possess the enzymes necessary for PTMs such as glycosylation, phosphorylation, or disulfide bond formation. These modifications are critical for the stability, activity, and function of many eukaryotic proteins. For proteins requiring PTMs, mammalian cell expression systems are often preferred despite their higher costs and complexity. |
Bacterial expression systems have transformed the landscape of molecular biology and biotechnology, providing a reliable and efficient means for producing recombinant proteins. From their foundational role in research to their impact on medicine, industry, and synthetic biology, their contributions are vast and diverse.
At Creative Biostructure, our bacterial protein expression services are tailored to meet your research and industrial needs. With advanced technology, expert optimization, and seamless scalability, we ensure the rapid production of high-quality proteins for multiple applications. Contact us today and enhance ate your structural biology projects with reliable, scalable bacterial expression services.
References
- Chen R. Bacterial expression systems for recombinant protein production: E. coli and beyond. Biotechnology Advances. 2012;30(5):1102-1107.
- Rosano GL, Ceccarelli EA. Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol. 2014;5.