Structural Research of P-type ATPase
P-type ATPase is present in all areas of life and constitutes a variety of cation transport proteins. It uses the energy generated by ATP hydrolysis to drive intramolecular motions that drive vectorial transport of ions, mainly H+, Na+, K+, Ca2+, and transition metal ions. In recent years, researchers have determined the structures of natural and recombinant P-type ATPase complexes based on the feasibility of X-ray free-electron-laser-based serial femtosecond crystallography (SFX). It is essential for understanding the function and molecular mechanism of P-type ATPase.
Mechanism of the P-type ATPase ion pump
Under conditions of high ATP: ADP ratios, the pump cycles directionally between the E1 and E2 states. In the E1 state, the pump has a high affinity for the exported substrate and the ion-binding site is open to cytoplasmic solutes. Autophosphorylation drives the formation of the ion-closed E1P-ADP state and a large structural rearrangement leads to the E2P state, which is now open to the other side. Dephosphorylation to E2, which is coupled to the binding and blocking of the imported substrate in the P2 ATPase. Subsequently, the pump is converted to E1 with the release of antagonistic ions on the cytoplasmic side, thus the pump is ready for another translocation cycle.
Structural analysis of P-type ATPases
All P-type ATPases have three cytoplasmic structural domains, A (agonist), P (phosphorylated), and N (nucleotide-bound), and a transmembrane (TM) structural domain with at least six transmembrane helices. Precise coupling between the cytoplasmic structural domain and the transmembrane helices is required, and therefore the topology is conserved throughout the family. Transmembrane fragments M1-M6 form the core transporter structural domain, which contains the ion-binding site. Together with the cytoplasmic structural domains, the transporter domains form the smallest functional unit. In addition to the P1 subfamily, P-type ATPases have additional supporting structural domains of transmembrane fragments M7-M10.
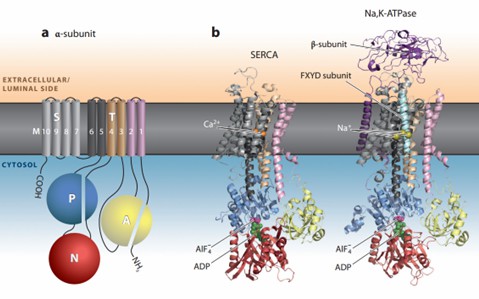 Figure 1. Structural architecture of P-type ATPases. (Dyla M, et al., 2020)
Figure 1. Structural architecture of P-type ATPases. (Dyla M, et al., 2020)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| H+, K+-ATPase with bound BeF and SCH28080 | Sus scrofa | Cryo-EM single particle analysis | 7 Å | 2XZB |
| H+, K+ - ATPase in complex with BYK99 | Sus scrofa | Cryo-EM single particle analysis | 6.7 Å | 5Y0B |
| H+, K+-ATPase with bound rubidium | Sus scrofa | Cryo-EM single particle analysis | 8 Å | 2YN9 |
| Na+, K+-ATPase in the E2.2K+ state | Squalus acanthias | Cryo-EM single particle analysis | 3.3 Å | 7Y45 |
| Na+, K+-ATPase in the E2P state formed by ATP with ouabain | Squalus acanthias | Cryo-EM single particle analysis | 3.4 Å | 7WYZ |
| Sodium-potassium pump in the E2. 2K+.Pi state | Squalus acanthias | X-ray diffraction | 2.4 Å | 2ZXE |
| Rb+-bound E2-MgF state of the gastric proton pump (Wild-type) | Sus scrofa | X-ray diffraction | 4.3 Å | 6JXK |
| Proton pump complexed with tegoprazan | Sus scrofa | X-ray diffraction | 3 Å | 7W47 |
| SR Ca2+-ATPase in the HnE2 state complexed with the Thapsigargin derivative DTB | Oryctolagus cuniculus | X-ray diffraction | 2.65 Å | 3NAL |
| Cadmium translocating P-type ATPase | Sulfitobacter sp. NAS-14.1 | X-ray diffraction | 3.25 Å | 7QBZ |
| Cadmium translocating P-type ATPase | Sulfitobacter sp. NAS-14.1 | X-ray diffraction | 3.11 Å | 7QC0 |
| KdpFABC complex in an E1 outward-facing state (state 1) | Escherichia coli K-12 | Cryo-EM single particle analysis | 3.7 Å | 6HRA |
| KdpFABC complex in an E2 inward-facing state (state 2) | Escherichia coli K-12 | Cryo-EM single particle analysis | 4 Å | 6HRB |
| Sodium-potassium pump with bound potassium and ouabain | Squalus acanthias | X-ray diffraction | 2.8 Å | 3A3Y |
| P5B-ATPase E2PiAlF/SPM | Thermochaetoides thermophila DSM 1495 | Cryo-EM single particle analysis | 3.7 Å | 7OP1 |
| P5B-ATPase E2P | Thermochaetoides thermophila DSM 1495 | Cryo-EM single particle analysis | 3.7 Å | 7OP5 |
| P5B-ATPase E2Pinhibit | Thermochaetoides thermophila DSM 1495 | Cryo-EM single particle analysis | 3.5 Å | 7OP8 |
| Proton ATPase | Neurospora crassa | Cryo-EM single particle analysis | 8 Å | 1MHS |
| A zinc-transporting PIB-type ATPase in E2P state | Shigella sonnei | X-ray diffraction | 2.705 Å | 4UMW |
| Ca2+-ATPas with G4 insertion | Listeria monocytogenes | X-ray diffraction | 3 Å | 6ZHH |
| Ion uptake in copper-transporting P1B-type ATPases | Archaeoglobus fulgidus | X-ray diffraction | 4.01 Å | 7R0G |
| Mg-ATPase Nucleotide binding domain | Escherichia coli K-12 | X-ray diffraction | 1.6 Å | 3GWI |
| Ion uptake in copper-transporting P1B-type ATPases | Archaeoglobus fulgidus | X-ray diffraction | 3.31 Å | 7R0H |
Table 1. Structural research of P-type ATPase.
As a distinguished service provider in the field of structural biology, Creative Biostructure offers a comprehensive range of services for the research of P-type ATPase and other biomolecules. With experienced experts and cutting-edge technologies, we are committed to providing one-stop structural analysis services to academia and the biotechnology industry.
Our team of expert scientists utilizes advanced techniques such as cryo-electron microscopy (cryo-EM), NMR spectroscopy, and X-ray crystallography to obtain high-resolution structural data. Contact us to learn more about how our capabilities can empower your research and propel you closer to achieving your scientific goals.
References
- Dyla M, et al. Structure and Mechanism of P-Type ATPase Ion Pumps. Annu Rev Biochem. 2020。89:583-603.
- Bublitz M, et al. Structural studies of P-type ATPase-ligand complexes using an X-ray free-electron laser. IUCrJ. 2015.2(Pt 4):409-420.
- Dyla M, et al. Structural dynamics of P-type ATPase ion pumps. Biochem Soc Trans. 2019。47(5):1247-1257.