Structural Research of F-type ATPase
There are four main types of ATPases: F-ATPase, V-ATPase, A-ATPase, and P-ATPase. The F/V/A-ATPase family is widely distributed in archaea, eubacteria, and eukaryotes. F-type ATPase mainly plays the role of ATP synthase and plays a key role in energy metabolism. Using ADP and inorganic phosphate as substrates, they exist on the plasma membrane of bacteria, the inner mitochondrial membrane, and the thylakoid membrane of chloroplasts.
F-ATPase is composed of two structural domains: the F0 domain extends to the mitochondrial membrane, which is a component of the membrane and consists of three different types of constituent proteins, namely a, b, and c. The F1 domain extends to the lumen, which is peripheral (on the side of the membrane where protons move in). F1 consists of five peptide units that bind to the surface of the F0 domain α three β three γδε Composition. The F1 domain is responsible for ATP turnover, while the F0 domain is responsible for ion translocation. In the cellular environment, F-ATPase typically acts as an ATP synthase rather than an ATPase. That is to say, it usually produces ATP from the proton gradient, rather than working in the other direction as V-ATPase usually does. They occasionally recover to ATPase in bacteria.
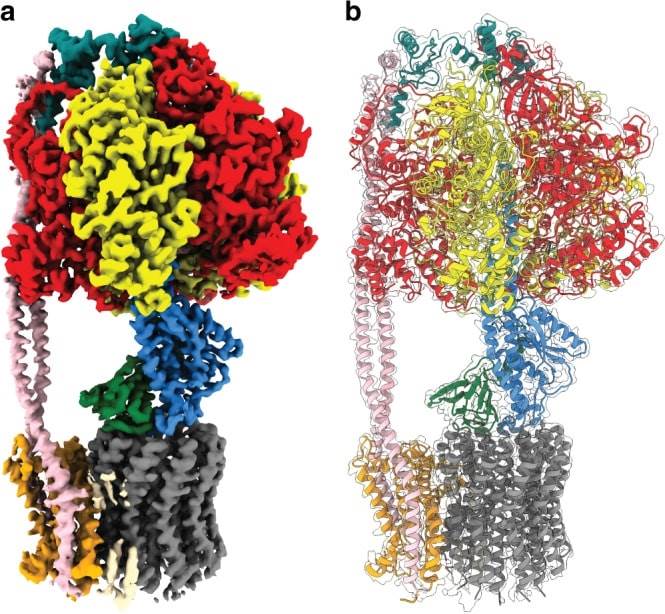 Figure 1. Cryo-EM map and atomic model of E. coli F1Fo ATP synthase. (Sobti M, et al., 2020)
Figure 1. Cryo-EM map and atomic model of E. coli F1Fo ATP synthase. (Sobti M, et al., 2020)
| Protein | Organism | Method | Resolution | PDB Entry ID |
|---|---|---|---|---|
| F1Fo ATP synthase, F1-peripheral stalk domain, state 1: Bos taurus | Bos taurus | Cryo-EM single particle analysis | 3.23 Å | 6YY0 |
| F1Fo-ATPase dimer, state1:state1: Bos taurus | Bos taurus | Cryo-EM single particle analysis | 9.20 Å | 7AJB |
| F1Fo synthase, ADP state 1A: Escherichia coli | Escherichia coli | Cryo-EM single particle analysis | 3.10 Å | 6OQR |
| ATP synthase (Fo) in nanodisc with inhibitor Bedaquiline: Saccharomyces cerevisiae | Saccharomyces cerevisiae, Saccharomyces cerevisiae S288C | X-ray diffraction | 4.20 Å | 6WTD |
| F1Fo ATP synthase in multiple states, State 1 catalytic (a) without exogenous ATP, backbone model: Saccharomyces cerevisiae | Saccharomyces cerevisiae | X-ray diffraction | 3.80 Å | 7TJY |
| F1Fo ATP synthase, R1, CF1FO: Spinacia oleracea | Spinacia oleracea | Cryo-EM single particle analysis | 3.35 Å | 6VON |
| F1Fo-ATP synthase, state 1: Acinetobacter baumannii | Acinetobacter baumannii ATCC 17978 | Cryo-EM single particle analysis | 3.10 Å | 7P2Y |
| F1Fo ATP synthase hexamer, composite model: Toxoplasma gondii | Toxoplasma gondii GT1 | Cryo-EM single particle analysis | 4.80 Å | 6TML |
Table 1. Structural research of F-type ATPase.
By analyzing the structure of F-type ATPase, we can gain a deeper understanding of its catalytic mechanism for ATP synthesis or decomposition and reveal how it utilizes proton gradients to drive this process. Due to the crucial role played by F-type ATPase in many diseases, understanding its structure can help design drugs targeting F-type ATPase and develop new treatment strategies. Meanwhile, F-type ATPase is one of the key enzymes in energy metabolism in organisms, and its structural analysis helps us understand the basic principles of bioenergy conversion and cellular metabolism.
In recent years, significant breakthroughs have been made in the structural research of F-type ATPase, and cryo-electron microscopy structural analysis allows us to gain a deeper understanding of how E. coli F1Fo ATP synthase adapts to symmetric mismatches. By using cryo-EM technology, the structure of F-type ATPase in its natural state can be observed, revealing its localization and assembly on the cell membrane.
Creative Biostructure is a scientific research service institution focusing on the field of protein structure biology. We provide a wide range of F-type ATPase structure research services for researchers, including but not limited to F-type ATPase protein expression and purification services, protein crystallization screening and optimization, and F-type ATPase structure analysis. If you are interested in exploring the structural research of F-type ATPases and would like to learn more about our services, please feel free to contact us at any time. Our team is always available to discuss your research needs and provide the best solution for your project.
Reference
- Sobti M, et al. Cryo-EM structures provide insight into how E. coli F1Fo ATP synthase accommodates symmetry mismatch. Nature Communications. 2020. 11(1).