Structural Research of Electron Transport Chain Complex II
Electron transport chain (ETC) is a critical process for energy production in living cells. It is composed of a series of protein complexes that transfer electrons from electron donors to electron acceptors, generating a proton gradient that drives ATP synthesis. ETC complex II, also known as succinate dehydrogenase (SQR) or fumarate reductase (QFR), is an essential component of the ETC, responsible for transferring electrons from succinate to ubiquinone. ETC complex II is an enzyme consisting of four subunits: SdhA, SdhB, SdhC, and SdhD. SdhA and SdhB form the catalytic core of the enzyme, while SdhC and SdhD anchor the enzyme to the inner mitochondrial membrane. Recent studies have used various techniques to reveal the structure and function of ETC complex II, including cryo-electron microscopy (cryo-EM) and X-ray crystallography.
Recent research has led to the discovery of a new class of respiratory complex II known as Mycobacterial Sdh1, which has an unknown electron transfer mechanism. Cryo-EM was used to determine the M. smegmatis Sdh1 structures alone and in the presence of ubiquinone-1, revealing its unique features. The enzyme comprises three subunits, with SdhC being the membrane-spanning subunit. Notably, a rarely observed Rieske-type [2Fe-2S] cluster was identified in the transmembrane region, which was found to be essential for quinone reduction activity. The study also showed that Mycobacterial Sdh1 has a novel electron transfer pathway and a unique substrate-binding site. The structures provide valuable information that could aid in the development of new antituberculosis drugs targeting this complex, given its essential role in Mycobacteria.
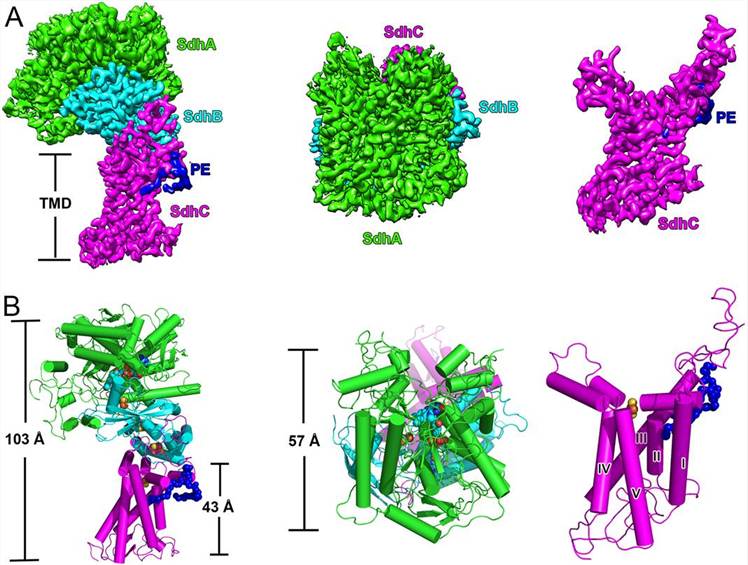 Figure 1. Overall structure of M. smegmatis Sdh1. (Zhou X, et al., 2021)
Figure 1. Overall structure of M. smegmatis Sdh1. (Zhou X, et al., 2021)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Native Fumarate Reductase Complex (expressed in E. coli) | Escherichia coli | X-ray diffraction | 3.30 Å | 1L0V |
| Fumarate Reductase Complex FrdA, E245Q mutant (expressed in E. coli) | Escherichia coli | X-ray diffraction | 4.22 Å | 5VPN |
| Fumarate Reductase Complex | Wolinella succinogenes | X-ray diffraction | 1.78 Å | 2BS2 |
| Formate dehydrogenase-N | Escherichia coli | X-ray diffraction | 1.60 Å | 1KQF |
| Succinate:quinone oxidoreductase (SQR, Complex II) (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.60 Å | 1NEK |
| Succinate:quinone oxidoreductase (SQR, Complex II) with Atpenin A5 (expressed in E. coli) | Escherichia coli | X-ray diffraction | 3.10 Å | 2ACZ |
| Succinate:quinone oxidoreductase (SQR, Complex II) with carboxin in Q-site (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.40 Å | 2WDQ |
| Succinate:quinone oxidoreductase (SQR, Complex II), H207T SdhB mutant (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.70 Å | 2WP9 |
| Succinate:quinone oxidoreductase (SQR, Complex II) (expressed in E. coli) | Escherichia coli | Cryo-EM single particle analysis | 3.60 Å | 6WU6 |
| Succinate:ubiquinone oxidoreductase (SQR, Complex II; pig heart) | Sus scrofa | X-ray diffraction | 2.40 Å | 1ZOY |
| Succinate:ubiquinone oxidoreductase (SQR, Complex II; chicken heart) w. carboxin inhibitor | Gallus gallus | X-ray diffraction | 2.06 Å | 2FBW |
| Succinate:ubiquinone oxidoreductase (SQR, Complex II; chicken heart) with TEO at the active site | Gallus gallus | X-ray diffraction | 1.74 Å | 2H88 |
| Succinate dehydrogenase Sdh1 complex in the apo form | Mycolicibacterium smegmatis | Cryo-EM single particle analysis | 2.88 Å | 7D6X |
Table 1. Structural Research of Electron Transport Chain Complex II.
Creative Biostructure is a top provider of services that analyze the structure of membrane proteins, which includes complex II. Our team of experts utilizes the advanced technology, such as nuclear magnetic resonance (NMR) spectroscopy, X-ray crystallography, and cryo-EM, to identify the structure and function of these proteins.
We have an experienced team of structural biologists who are dedicated to providing high-quality analysis services. We collaborate closely with our clients to ensure that their research goals are met, and we provide comprehensive reports that thoroughly explain our research findings.
If you are interested in learning more about our services for analyzing membrane protein structures, please do not hesitate to contact us. Our team of experts would be delighted to discuss your project with you and provide you with a no-cost quote. Allow us to assist you in advancing your research and bringing your discoveries to fruition.
References
- Su C C, et al. A 'Build and Retrieve' methodology to simultaneously solve cryo-EM structures of membrane proteins. Nature Methods. 2021, 18(1): 69-75.
- Zhou X, et al. Architecture of the mycobacterial succinate dehydrogenase with a membrane-embedded Rieske FeS cluster. Proceedings of the National Academy of Sciences. 2021, 118(15): e2022308118.