Structural Research of Calcium Ion-Selective Channels
The regulation of calcium ion concentrations within cells is tightly controlled by a variety of mechanisms, including the activity of calcium ion-selective channels. These channels are responsible for allowing calcium ions to flow across cellular membranes, and their malfunction has been implicated in a number of diseases, including heart disease, Alzheimer's disease, and cancer.
One of the most well-studied types of calcium ion-selective channels is the voltage-gated calcium (Cav) channel, which is found in a variety of cell types throughout the body. Several high-resolution structures of Cav channel have been obtained using X-ray crystallography and cryo-electron microscopy (cryo-EM) techniques. These structures have revealed the location of important functional domains within the channel, as well as the specific interactions between the channel and various ligands, such as calcium ions and drugs that modulate channel activity.
The Cav1 channels Cav1 channels are hetero-multimeric complexes encompassing various subunits including the pore-forming α1 core subunit and auxiliary subunits α2δ, β, and γ. The α1 subunit folds into four homologous repeats I-IV, which are aligned in a canonical voltage-gated ion channel fold. Each repeat encompasses six transmembrane segments, denoted as S1-S6, which are arranged in two functional entities. The peripheral voltage sensing domain of the Cav1 channels comprises the S1-S4 segments in each repeat. The S5 and S6 segments, in combination with the intervening segments, including the selectivity filters and the supporting helices P1 and P2, derived from the four repeats, ultimately come together to form the central ion-conducting pore domain of these channels.
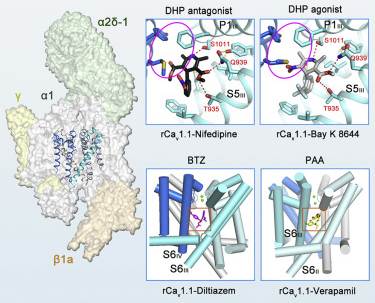 Figure 1. Molecular basis for rCav1.1 modulation by chemical ligands. (Zhao Y, et al., 2019)
Figure 1. Molecular basis for rCav1.1 modulation by chemical ligands. (Zhao Y, et al., 2019)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Ca2+ selectivity of a voltage-gated calcium channel (expressed in Trichoplusia ni) | Aliarcobacter butzleri | X-ray diffraction | 2.75 Å | 4MS2 |
| CavAb voltage-gated calcium channel (expressed in Trichoplusia ni) | Aliarcobacter butzleri | X-ray diffraction | 2.70 Å | 5KLB |
| Voltage-gated calcium channel Cav1.1 complex (expressed in E. coli) | Oryctolagus cuniculus | Cryo-EM single particle analysis | 4.20 Å | 3JBR |
| Voltage-gated calcium channel Cav1.1 complex (expressed in E. coli) | Oryctolagus cuniculus | Cryo-EM single particle analysis | 3.60 Å | 5GJV |
| Cav1.1-Nifedipine Complex (expressed in E. coli) | Oryctolagus cuniculus | Cryo-EM single particle analysis | 2.90 Å | 6JP5 |
| CaV1.1 voltage-gated calcium channel in nanodiscs, in presence of amlodipine | Oryctolagus cuniculus | Cryo-EM single particle analysis | 2.90 Å | 7JPX |
| L-type voltage-gated calcium channel Cav1.3 (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 3.00 Å | 7UHG |
| N-type voltage-gated calcium channel Cav2.2 (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 3.10 Å | 7MIY |
| N-type voltage gated calcium channel CaV2.2-alpha2/delta1-beta1 complex, apo state (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 2.80 Å | 7VFS |
| Cav2.3 R-type voltage-gated calcium channel, wild-type (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 3.10 Å | 8EPL |
| R-type voltage-gated CaV2.3-alpha2/delta1-beta1 channel complex in the ligand-free (apo) state (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 3.10 Å | 7XLQ |
| CaV3.1 voltage-gated calcium channel (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 3.30 Å | 6KZO |
| Calcium release-activated calcium (CRAC) channel ORAI (expressed in Komagataella pastoris) | Drosophila melanogaster | X-ray diffraction | 3.35 Å | 4HKR |
| CRAC channel Orai in an open conformation; H206A gain-of-function mutation (expressed in Komagataella pastoris) | Drosophila melanogaster | X-ray diffraction | 6.71 Å | 6BBF |
| Calcium release-activated calcium channel protein 1, P288L mutant (expressed in HEK293 cells) | Drosophila melanogaster | X-ray diffraction | 4.50 Å | 6AKI |
| CRAC channel Orai in an open conformation; H206A gain-of-function mutation in complex with an antibody (expressed in Komagataella pastoris) | Drosophila melanogaster | Cryo-EM single particle analysis | 3.30 Å | 7KR5 |
| Stromal interaction molecule 1 coiled-coil 1 fragment (expressed in E. coli) | Homo sapiens | Solution NMR | / | 6YEL |
| YetJ pH-sensitive calcium-leak channel, pH 8 (closed form) (expressed in E. coli) | Bacillus subtilis | X-ray diffraction | 1.95 Å | 4PGR |
| YetJ pH-sensitive calcium-leak channel (expressed in E. coli) | Bacillus subtilis | X-ray diffraction | 2.50 Å | 6NQ7 |
| RyR1 ryanodine receptor, closed state in complex with FKBP12 (expressed in E. coli) | Oryctolagus cuniculus | Cryo-EM single particle analysis | 3.80 Å | 3J8H |
| RyR1 (EGTA-only dataset, all particles) (expressed in E. coli) | Homo sapiens | Cryo-EM single particle analysis | 4.40 Å | 5TB0 |
| Ryanodine Receptor 1 Repeat12 Domain (expressed in E. coli) | Oryctolagus cuniculus | X-ray diffraction | 1.55 Å | 5C30 |
| Ryanodine Receptor 1 in nanodiscs in the presence of calcium and ATP | Oryctolagus cuniculus | Cryo-EM single particle analysis | 8.20 Å | 6FOO |
| RyR1 ryanodine receptor in complex with Ca2+ and chlorantraniliprole (CHL) (expressed in E. coli) | Oryctolagus cuniculus | Cryo-EM single particle analysis | 4.70 Å | 7CF9 |
| RyR1 ryanodine receptor embedded in a lipid bilayer, primed model (expressed in E. coli) | Oryctolagus cuniculus | Cryo-EM single particle analysis | 3.36 Å | 7M6A |
| RyR1 in the presence of AMP-PCP in nanodisc | Oryctolagus cuniculus | Cryo-EM single particle analysis | 4.30Å | 7K0T |
| Ryanodine Receptor type 1 mutant R164C in complex with FKBP12.6 (expressed in HEK293 cells) | Oryctolagus cuniculus | Cryo-EM single particle analysis | 3.54 Å | 6WOT |
| RyR1 disease mutant Y523S in complex with FKBP12.6 embedded in lipidic nanodisc in the closed state (expressed in HEK293 cells) | Oryctolagus cuniculus | Cryo-EM single particle analysis | 4.00 Å | 7T64 |
| RyR2 ryanodine receptor, closed state | Sus scrofa | Cryo-EM single particle analysis | 4.40 Å | 5GO9 |
| RyR2 ryanodine receptor bound to FKBP12.6 interacting with human calmodulin (CaM), apo CaM state (expressed in E. coli) | Sus scrofa | Cryo-EM single particle analysis | 3.60 Å | 6JI8 |
| RyR2 (Ca2+ alone dataset) | Sus scrofa | Cryo-EM single particle analysis | 6.10 Å | 6JG3 |
| RyR2 ryanodine receptor SPRY1 domain critical in binding FKBP12 (expressed in E. coli) | Mus musculus | X-ray diffraction | 1.21 Å | 5C33 |
| RyR2 ryanodine receptor, EGTA dataset, class 1&2, closed state (expressed in HEK293 cells) | Mus musculus | Cryo-EM single particle analysis | 3.30 Å | 7VML |
| InsP3R1 Inositol-1,4,5-trisphosphate receptor | Rattus norvegicus | Cryo-EM single particle analysis | 4.70 Å | 3JAV |
| InsP3R1 Inositol-1,4,5-trisphosphate receptor in lipid nanodisc, the apo-state | Rattus norvegicus | Cryo-EM single particle analysis | 3.30 Å | 7LHE |
| InsP3R1 Inositol-1,4,5-trisphosphate receptor in the presence of Calcium/IP3/ATP | Rattus norvegicus | Cryo-EM single particle analysis | 3.50 Å | 8EAR |
| InsP3R3 Inositol-1,4,5-trisphosphate receptor, apo state (expressed in Sf9 cells) | Homo sapiens | Cryo-EM single particle analysis | 3.49 Å | 6DQJ |
| InsP3R3 Inositol-1,4,5-trisphosphate receptor and presence of self-binding peptide (expressed in Spodoptera frugiperda) | Homo sapiens | Cryo-EM single particle analysis | 3.77 Å | 6UQK |
| IP3 and ATP bound type 3 IP3 receptor in the pre-active A state (expressed in Spodoptera frugiperda) | Homo sapiens | Cryo-EM single particle analysis | 3.20 Å | 7T3P |
| Mitochondrial calcium uniporter (MCU) (expressed in E. coli) | Caenorhabditis elegans | Solution NMR | / | 5ID3 |
| Mitochondrial calcium uniporter (MCU), full length (expressed in E. coli) | Neurospora crassa | Cryo-EM single particle analysis | 3.70 Å | 6DT0 |
| Mitochondrial calcium uniporter (MCU), full length (expressed in E. coli) | Aspergillus fischeri | Cryo-EM single particle analysis | 3.80 Å | 6D7W |
| Mitochondrial calcium uniporter (MCU), full length (expressed in E. coli) | Metarhizium acridum | X-ray diffraction | 3.10 Å | 6C5W |
| Mitochondrial calcium uniporter (MCU), full length (expressed in Komagataella pastoris) | Cyphellophora europaea | Cryo-EM single particle analysis | 3.20 Å | 6DNF |
| Mitochondrial calcium uniporter (MCU) in complex with EMRE (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 3.80 Å | 6O58 |
| Mitochondrial calcium uniporter holocomplex in low Ca2+ (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 3.20 Å | 6WDN |
| Mitochondrial calcium uniporter (MCU) holocomplex (uniplex) in nanodiscs, high calcium state (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 4.17 Å | 6XJV |
| Mitochondrial calcium uniporter (MCU) in complex with MICU1/MICU2 subunits (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 3.60 Å | 6K7Y |
| RyR2 ryanodine receptor, PKA phosphorylated in the closed state (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 3.11 Å | 7U9Q |
Table 1. Structural Research of Calcium Ion-Selective Channels.
At Creative Biostructure, we are committed to providing high-quality structural analysis services and expertise to help our clients advance their research and achieve their scientific goals. Our team of experienced scientists has extensive expertise in X-ray crystallography, cryo-EM, NMR spectroscopy and other structural biology techniques, and we use first-class equipment and software to obtain high-resolution structures of our client's target proteins.
In general, our structural analysis services include protein expression and purification, structure determination using X-ray crystallography or cryo-EM, and structure validation and refinement. We also offer additional services, such as ligand binding studies and functional assays, to provide a comprehensive understanding of the structure and function of target proteins.
If you are interested in learning more about our structural analysis services for calcium ion-selective channels or other membrane proteins, please don't hesitate to contact us. Our team of experts will be happy to discuss your project and provide a customized solution to meet your needs.
References
- Zhao Y, et al. Molecular basis for ligand modulation of a mammalian voltage-gated Ca2+ channel. Cell. 2019, 177(6): 1495-1506. e12.
- Gao Y, et al. Molecular insights into the gating mechanisms of voltage-gated calcium channel CaV2. 3. Nature Communications. 2023, 14(1): 516.
- Schmitz E A, Takahashi H, Karakas E. Structural basis for activation and gating of IP3 receptors. Nature Communications. 2022, 13(1): 1408.
