What Is Mass Spectrometry?
Mass spectrometry (MS) is an analytical technique used to measure the mass-to-charge ratio (m/z) of ions. It is employed to identify the composition of a sample by analyzing the mass of its molecules. MS is highly versatile and can be used to analyze a wide range of compounds, including small organic molecules, large proteins, and even complex biomolecules. The technique plays a crucial role in both qualitative and quantitative analysis, allowing scientists to determine the structure, molecular weight, and chemical composition of substances.
In mass spectrometry, samples are ionized, and the ions are then separated and detected based on their m/z ratios. This information is used to generate a mass spectrum, a graphical representation that displays the relative abundance of detected ions. Each peak on the mass spectrum corresponds to a fragment of the analyte, and the distribution of these peaks helps in identifying the chemical structure and molecular weight of the compound.
Mass spectrometry is a powerful tool in a variety of fields, including chemistry, biochemistry, pharmaceuticals, and structural biology. It is often used for molecular identification, isotope analysis, studying complex mixtures, and elucidating protein structures.
History of Mass Spectrometry
Mass spectrometry originated in the early 20th century when British physicist J.J. Thomson developed a method to measure the mass of charged particles, using it in 1913 to identify isotopes of neon. Thomson's technique, the "parabola method," separated ions by their mass-to-charge ratio using magnetic and electric fields. Later, Francis W. Aston improved upon Thomson's work by creating the first modern mass spectrometer, leading to the discovery of many naturally occurring isotopes. For his contributions, Aston received the Nobel Prize in Chemistry in 1922.
In the mid-20th century, technological advances, such as the development of vacuum systems, ionization techniques, and detection methods, revolutionized mass spectrometry. In particular, the introduction of electron ionization (EI) in 1953 by A.J. Dempster provided a more efficient means of ionizing molecules. This advancement, along with the development of quadrupole mass analyzers, time-of-flight (TOF) detectors, and tandem mass spectrometry (MS/MS), significantly enhanced the precision and sensitivity of the technique.
The most significant advancements in recent decades include the invention of electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI), both of which enabled the analysis of large biomolecules, such as proteins and nucleic acids. These breakthroughs have made mass spectrometry a cornerstone in the fields of proteomics, genomics, and structural biology.
Principles and Instrumentation of Mass Spectrometry
Mass spectrometry operates on the principle of ionizing molecules, separating the resulting ions based on their mass-to-charge ratio, and detecting them to generate a mass spectrum. The key components of a mass spectrometer include the ion source, the mass analyzer, and the detector.
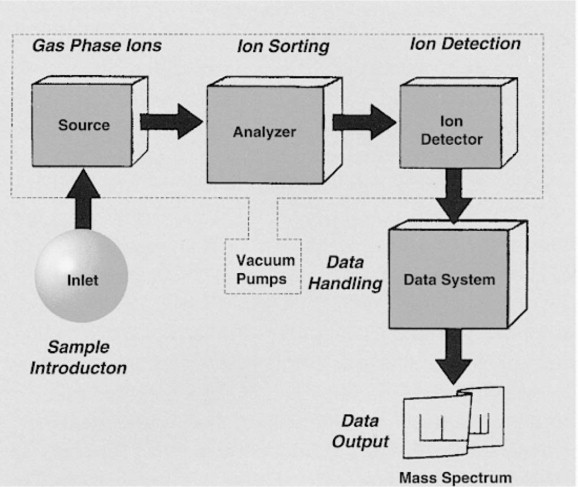 Figure 1. The components of a mass spectrometer (Zagorevskii, 2003).
Figure 1. The components of a mass spectrometer (Zagorevskii, 2003).
Ion Source
The first step in mass spectrometry is ionization, where the sample is converted into charged particles. This can be achieved through various ionization techniques, depending on the sample's properties. Common ionization methods include:
- Electron Ionization (EI): Often used for small, volatile molecules, EI involves bombarding the sample with a stream of electrons, causing it to lose electrons and form positively charged ions.
- Electrospray Ionization (ESI): ESI is used for large biomolecules like proteins. It involves spraying the sample through a high-voltage needle, creating charged droplets that evaporate to leave charged ions in the gas phase.
- Matrix-Assisted Laser Desorption/Ionization (MALDI): In MALDI, the sample is mixed with a matrix material and irradiated with a laser. The laser energy is absorbed by the matrix, leading to the formation of ions from the analyte.
Mass Analyzer
Once the sample is ionized, the ions are directed into the mass analyzer, which separates them based on their m/z ratio. The most common types of mass analyzers include:
- Quadrupole Mass Analyzer: This device uses four parallel rods to create oscillating electric fields that selectively allow ions of specific m/z ratios to pass through, while others are filtered out.
- Ion Trap: Ion traps confine ions in a small space using electric and magnetic fields. The ions are then sequentially released and detected based on their m/z ratios.
- Orbitrap: The Orbitrap analyzer traps ions in an electrostatic field and measures their oscillation frequencies, which are proportional to the m/z ratio.
- Time-of-Flight (TOF) Analyzer: In TOF analyzers, ions are accelerated by an electric field, and their flight time to the detector is measured. Since ions with lower mass travel faster, this method allows for the precise determination of their m/z ratio.
Detector
After the ions are separated by the mass analyzer, they are directed to the detector, which records the number and intensity of ions at each m/z ratio. Common detectors include electron multipliers and microchannel plates, which convert ion impacts into electrical signals that are processed to generate the mass spectrum.
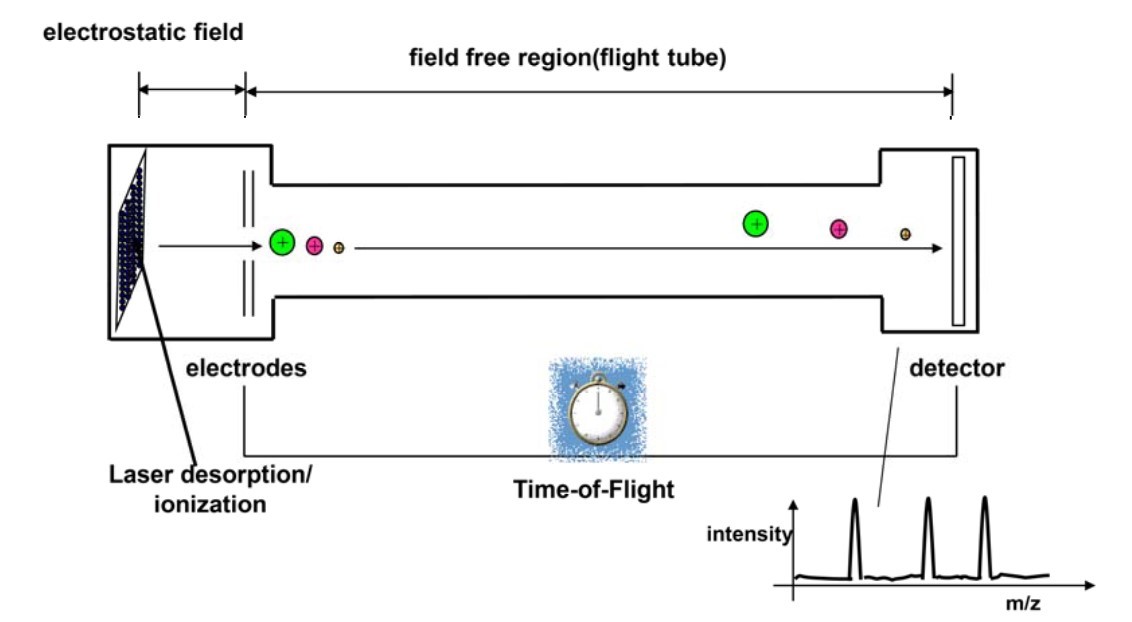 Figure 2. MALDI analyses with a time-of-flight mass analyzer (Pavlovic et al., 2013).
Figure 2. MALDI analyses with a time-of-flight mass analyzer (Pavlovic et al., 2013).
Separation Techniques Combined with Mass Spectrometry
Mass spectrometry is often combined with various separation techniques to improve the resolution and analysis of complex mixtures. The most common combinations include:
Gas Chromatography-Mass Spectrometry (GC-MS): In GC-MS, gas chromatography is used to separate volatile compounds in a sample based on their boiling points before they are introduced into the mass spectrometer. This combination is ideal for the analysis of small organic molecules, environmental pollutants, and complex mixtures such as essential oils or petrochemicals.
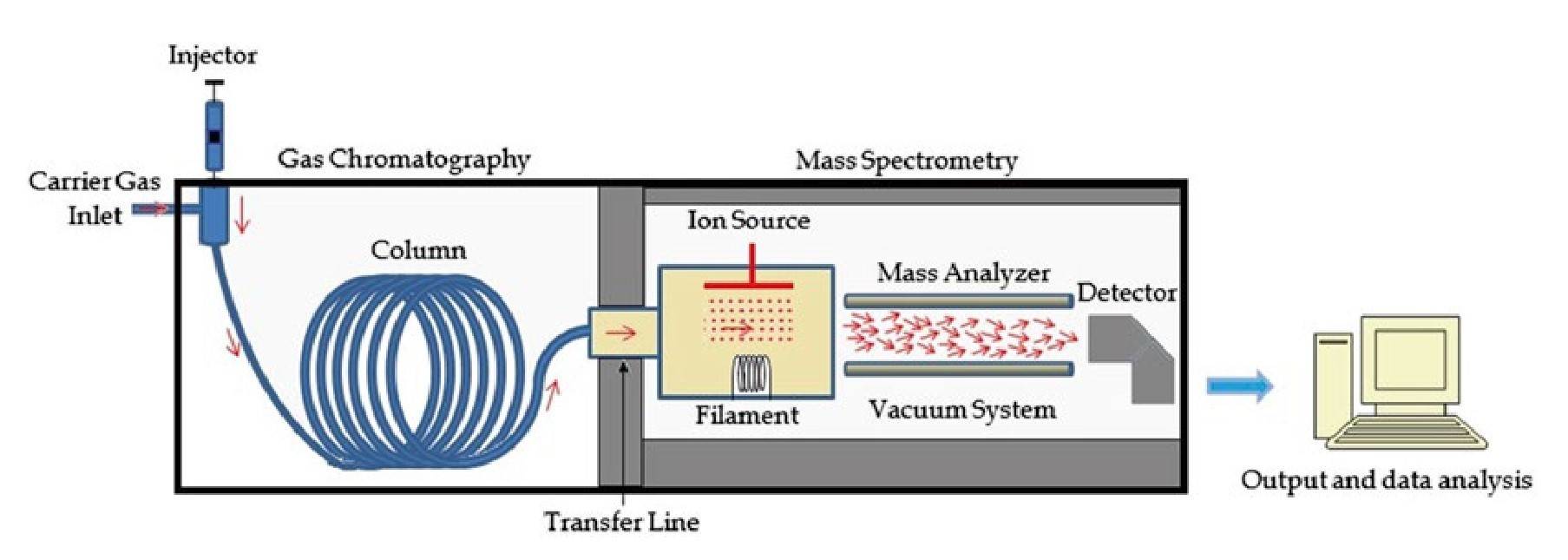 Figure 3 Diagram of a GC-MS system (Emwas et al., 2015).
Figure 3 Diagram of a GC-MS system (Emwas et al., 2015).
Liquid Chromatography-Mass Spectrometry (LC-MS): LC-MS is used to separate non-volatile compounds, including proteins, peptides, and pharmaceuticals, in liquid form before mass spectrometric analysis. High-performance liquid chromatography (HPLC) is commonly paired with MS for the analysis of complex biological samples and drug metabolites.
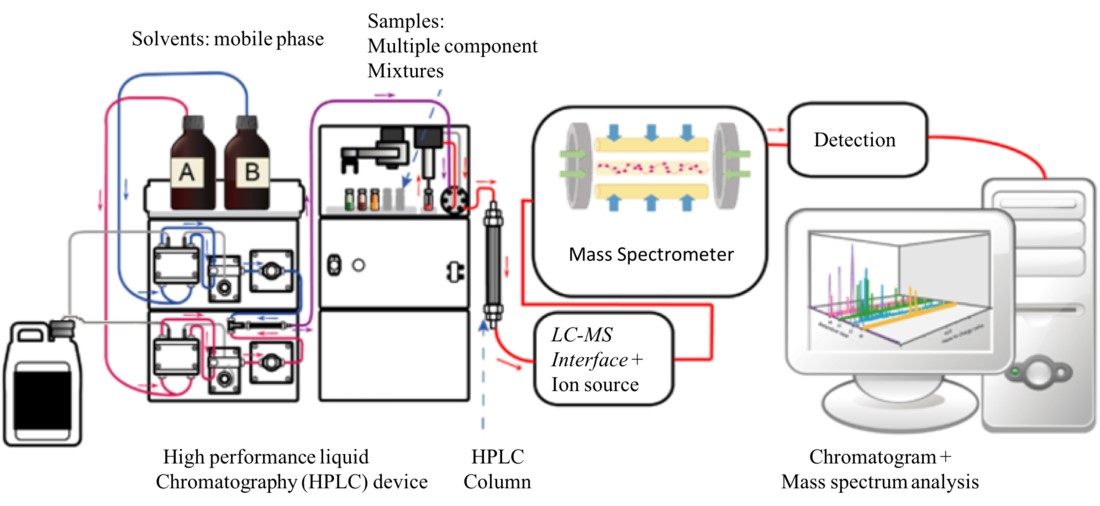 Figure 4. Diagram of an LC-MS system.
Figure 4. Diagram of an LC-MS system.
Capillary Electrophoresis-Mass Spectrometry (CE-MS): CE-MS is a powerful technique for separating charged species, such as peptides and nucleotides, based on their size and charge in an electric field. This method is particularly useful in proteomics and the analysis of biomolecules.
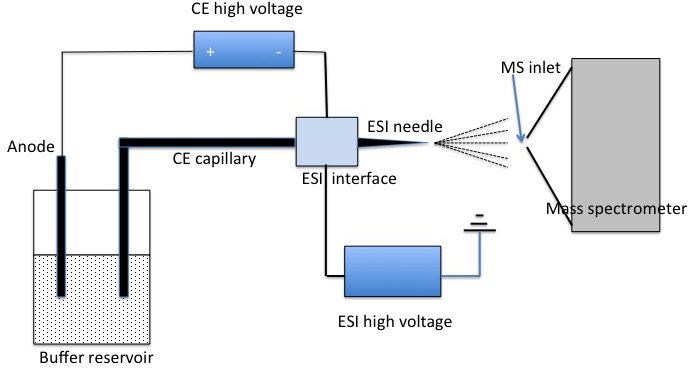 Figure 5. Diagram of a CE-MS system.
Figure 5. Diagram of a CE-MS system.
Applications of Mass Spectrometry
Mass spectrometry has become an indispensable tool in structural biology, providing critical insights into the structure, dynamics, and interactions of biomolecules such as proteins, nucleic acids, and lipids. Some key applications in structural biology include:
Protein Identification and Characterization: MS is widely used in proteomics to identify and characterize proteins by analyzing their peptide fragments. Tandem mass spectrometry (MS/MS) allows for the sequencing of peptides, which can be mapped back to the corresponding protein sequences. This is crucial for understanding protein functions, post-translational modifications, and interactions.
Determination of Protein-Protein Interactions: Cross-linking mass spectrometry (XL-MS) involves chemically cross-linking proteins at interacting sites and then analyzing the cross-linked complexes by MS. This provides valuable information about protein-protein interaction sites and helps in modeling complex molecular structures.
Top-Down Proteomics: In top-down proteomics, intact proteins are analyzed directly by MS without prior digestion into peptides. This approach allows for the detailed study of protein isoforms, post-translational modifications, and higher-order structures. It is particularly useful for characterizing complex proteins such as antibodies.
Hydrogen-Deuterium Exchange (HDX-MS): HDX-MS is used to study protein folding and dynamics by measuring the exchange of hydrogen atoms in the protein backbone with deuterium in the solvent. By comparing the rates of exchange in different regions of the protein, scientists can infer information about the protein's secondary and tertiary structures.
Native Mass Spectrometry: Native MS allows for the analysis of biomolecules in their native, non-denatured states. This technique is used to study large protein complexes, nucleic acids, and membrane proteins in their functional forms, providing insights into their structure, stoichiometry, and interactions.
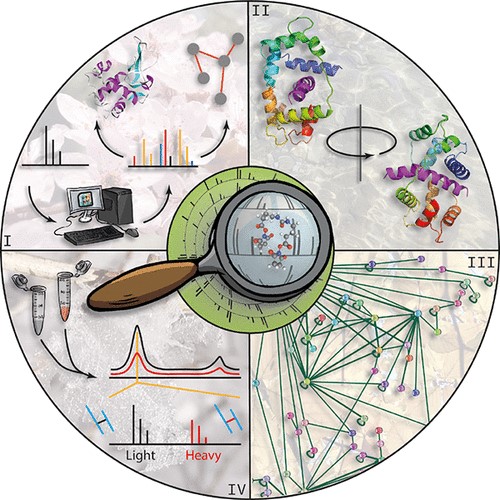 Figure 6. Applications of mass spectrometry. MS is used in combination with other techniques to study protein structure (I), protein dynamics (II), protein-protein interactions (III), and protein identification (IV) (Piersimoni et al., 2022).
Figure 6. Applications of mass spectrometry. MS is used in combination with other techniques to study protein structure (I), protein dynamics (II), protein-protein interactions (III), and protein identification (IV) (Piersimoni et al., 2022).
In summary, mass spectrometry is a versatile and powerful analytical tool that has evolved over the last century into a vital instrument for modern science, particularly in structural biology. Its ability to ionize, separate, and detect ions based on their mass-to-charge ratio allows for the detailed study of complex biomolecules, making it invaluable for applications such as protein identification, characterization, and interaction studies. When combined with separation techniques like gas and liquid chromatography, mass spectrometry provides unparalleled resolution and accuracy in the analysis of molecular structures, offering profound insights into the mechanisms that govern biological processes.
At Creative Biostructure, we offer comprehensive mass spectrometry (MS) analysis services, providing precise molecular identification and quantification for structural biology research. Our advanced MS technology, combined with complementary techniques like chromatography and electron microscopy, ensures thorough biomolecular characterization and enhanced accuracy in your studies. With a commitment to delivering high-quality results, we are your trusted partner in pushing the boundaries of scientific discovery. Contact us today to learn how our MS services can elevate your research.
References
- Comprehensive Coordination Chemistry II, Chapter Mass spectrometry, pp. 367–380, Elsevier, 2003. Zagorevskii, D.
- Metabonomics, Chapter Gas chromatography–mass spectrometry of biofluids and extracts, vol. 1277, pp. 91–112, edited by J. T. Bjerrum, Springer New York, 2015. Emwas, A.-H. M., Al-Talla, Z. A., Yang, Y., & Kharbatia, N. M.
- Pavlovic, M., Huber, I., Konrad, R., & Busch, U. (2013). Application of maldi-tof ms for the identification of food borne bacteria. The Open Microbiology Journal, 7(1), 135–141.
- Piersimoni, L., Kastritis, P. L., Arlt, C., & Sinz, A. (2022). Cross-linking mass spectrometry for investigating protein conformations and protein–protein interactions─a method for all seasons. Chemical Reviews, 122(8), 7500–7531.