Structural Research of Dehydrogenases
Dehydrogenase is an oxidoreductase. They catalyze the oxidation of substrates by transferring hydrogen (hydride) to receptors (or coenzymes). Dehydrogenases are classified based on donor groups (such as alcohols or aldehydes) and receptors. Receptors include nicotinamide adenine dinucleotide (NAD+or NADH), nicotinamide adenine dinucleotide phosphate (NADP+or NADPH), flavin adenine dinucleotide (FAD), flavin mononucleotide (FMN), and cytochrome. Dehydrogenases are important in various physiological processes, including normal cellular metabolism as part of the TCA (Krebs) cycle or cancer metabolism in the glycolytic pathway.
Non proton pumped type II NADH dehydrogenase (NDH-2) is a small unit membrane protein (40-60 kDa) that plays a core role in bacterial respiratory metabolism and mitochondria in fungi, plants, and protozoa. The crystal structure of bacterial NDH-2 enzyme from Caldalkalibacillus thermarum can be elucidated by X-ray crystallography with a resolution of 2.5Å. The NDH-2 structure reveals a homologous dimer organization with a unique dimer interface. NDH-2 is located on the cell plasma membrane through two separated C-terminal membrane anchoring regions, which are crucial for membrane localization and FAD binding. The overall structure of bacterial NDH-2 homodimer is shown in Figure 1. The first (cyan residues 2-109 and 263-345) and the second (gold residues 110-262) Rossmann folding domain and C-terminal membrane anchoring domain (magenta 346-398) are shown as strip plots. The noncovalent binding FAD is shown as a space filled model. In the left panel, the molecules are displayed from the side facing the membrane, while in the right panel, the molecules are rotated 90° and viewed in the "side" direction, and the magenta membrane anchor region is visible at the top.
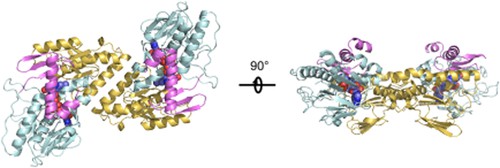 Figure 1. Overall structure of the bacterial NDH-2 homodimer. (HEIKAL A, et al., 2014)
Figure 1. Overall structure of the bacterial NDH-2 homodimer. (HEIKAL A, et al., 2014)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Glycerol-3-phosphate dehydrogenase (GlpD, native) | Escherichia coli | X-ray diffraction | 1.75 Å | 2QCU |
| Glycerol-3-phosphate dehydrogenase (SeMet-GlpD) | Escherichia coli | X-ray diffraction | 1.95 Å | 2R4J |
| Glycerol-3-phosphate dehydrogenase (GlpD-2-PGA) | Escherichia coli | X-ray diffraction | 2.3 Å | 2R45 |
| Glycerol-3-phosphate dehydrogenase (GlpD-PEP) | Escherichia coli | X-ray diffraction | 2.10 Å | 2R46 |
| Glycerol-3-phosphate dehydrogenase (GlpD-DHAP) | Escherichia coli | X-ray diffraction | 2.10 Å | 2R4E |
| PutA proline utilization protein | Geobacter sulfurreducens PCA | X-ray diffraction | 1.90 Å | 4NM9 |
| PutA proline utilization protein in complex with L-tetrahydro-2-furoic acid | Geobacter sulfurreducens PCA | X-ray diffraction | 2.10 Å | 4NMA |
| PutA proline utilization protein in complex with L-lactate | Geobacter sulfurreducens PCA | X-ray diffraction | 2.20 Å | 4NMB |
| PutA proline utilization protein in complex with Zwittergent 3-12 | Geobacter sulfurreducens PCA | X-ray diffraction | 1.90 Å | 4NMC |
| PutA proline utilization protein reduced with dithionite | Geobacter sulfurreducens PCA | X-ray diffraction | 1.98 Å | 4NMD |
| PutA proline utilization protein inactivated by N-propargylglycine | Geobacter sulfurreducens PCA | X-ray diffraction | 2.09 Å | 4NME |
| PutA proline utilization protein inactivated by N-propargylglycine and complexed with menadione bisulfite | Geobacter sulfurreducens PCA | X-ray diffraction | 1.95 Å | 4NMF |
| Ndi1 NADH Dehydrogenase | Saccharomyces cerevisiae S288C | X-ray diffraction | 2.70 Å | 4G9K |
| Ndi1-NAD+ complex | Saccharomyces cerevisiae S288C | X-ray diffraction | 2.90 Å | 4GAP |
| Ndi1-UQ2 complex | Saccharomyces cerevisiae S288C | X-ray diffraction | 3.00 Å | 4GAV |
| Ndi1 NADH Dehydrogenase, apo form (expressed in E. coli) | Saccharomyces cerevisiae S288C | X-ray diffraction | 2.39 Å | 4G6G |
| Ndi1 NADH Dehydrogenase, with NADH | Saccharomyces cerevisiae S288C | X-ray diffraction | 2.26 Å | 4G6H |
| Ndi1 NADH Dehydrogenase, with Quinone | Saccharomyces cerevisiae S288C | X-ray diffraction | 2.48 Å | 4G74 |
| Ndi1 NADH Dehydrogenase, with NAH and Quinone | Saccharomyces cerevisiae S288C | X-ray diffraction | 2.52 Å | 4G73 |
| NDH-2 NADH dehydrogenase (expressed in E. coli) | Saccharomyces cerevisiae S288C | X-ray diffraction | 2.50 Å | 4NWZ |
| (S)-Mandelate dehydrogenase (MDH) | Saccharomyces cerevisiae S288C | X-ray diffraction | 2.20 Å | 6BFG |
Table 1. Structural Research of Dehydrogenases.
Creative Biostructure's X-ray crystallography technology utilizes state-of-the-art instrumentation and experienced laboratory personnel to provide high-quality 3D structural analysis of membrane proteins. We analyze and process samples to obtain high-resolution crystallographic images of membrane proteins, which reveal the internal structure and localization of functional regions. If you are interested in learning more about our protein structural analysis services, please contact us for more information.
References
- YEH J I, et al. Structure of glycerol-3-phosphate dehydrogenase, an essential monotopic membrane enzyme involved in respiration and metabolism. Proceedings of the National Academy of Sciences, 2008, 105(9): 3280–3285.
- SINGH H, et al. Structures of the puta peripheral membrane flavoenzyme reveal a dynamic substrate-channeling tunnel and the quinone-binding site. Proceedings of the National Academy of Sciences, 2014, 111(9): 3389–3394.
- IWATA M, et al. The structure of the yeast NADH dehydrogenase (NDI1) reveals overlapping binding sites for water- and lipid-soluble substrates. Proceedings of the National Academy of Sciences, 2012, 109(38): 15247–15252.
- FENG Y, et al. Structural insight into the type-II mitochondrial NADH dehydrogenases. Nature, 2012, 491(7424): 478–482.
- HEIKAL A, et al. Structure of the bacterial type II NADH dehydrogenase: A monotopic membrane protein with an essential role in energy generation. Molecular Microbiology, 2014, 91(5): 950–964.
- SUKUMAR N, et al. Structure of the monotopic membrane protein (s)-mandelate dehydrogenase at 2.2 Å resolution. Biochimie, 2018, 154: 45–54.