Structural Research of Solute Sodium Symporter (SSS) Family
The solute sodium symporter (SSS) family consists of integral membrane proteins that use sodium gradients to drive the transport of solutes such as sugars, amino acids, vitamins, nucleosides, inositol, or ions across membranes. The SSS family members have been identified in bacteria, archaea, and eukaryotes. For example, the human sodium/iodine symporter protein (NIS) and sodium/glucose symporter protein (SGLT1) have been associated with diseases such as defective iodine transport or glucose-galactose malabsorption; and the bacterial sodium/proline symporter PutP and sodium/sialic acid symporter SiaT play essential roles in bacterial-host interactions.
Advances in research on SSS family proteins
Understanding the molecular mechanisms of transporter protein function requires detailed structural information. Indeed, X-ray crystallography has made significant progress in understanding channel-type proteins and ion pumps. Most of the experimental evidence for topological arrangements comes from research on the E. coli Na+ /proline symporter protein PutP. Circular dichroism measurements of purified and reconstituted PutP and the use of fixed-point spin labeling and electron paramagnetic resonance (EPR) spectroscopy are used to test the altered structural arrangement of PutP. Information on the spin-labeling morphology is obtained by analyzing the residual mobility of site-specifically linked nitroxide side chains and determining the collision frequencies of nitroxide with nonpolar oxygen and polar CROX.
Structural analysis of SSS family members
SSS proteins have 13 to 15 transmembrane helices (TMS) and usually contain 13 TMS cores, but the number of TMS varies among family members. For instance, PutP has a 13 TMS topology in the periplasmic N-terminus and cytoplasmic C-terminus. Residues essential for substrate and Na+ binding in PutP are present in TMS 2, 7, and 9 and adjacent loops. The researchers report the crystal structure of the SSS family member sodium: galactose homotransporter vSGLT. The approximately 3.0 Å structure contains 14 transmembrane α-helices oriented in the inward-facing conformation, and its core structure consists of an inverted repeat sequence of five TM helices (TM2 to TM6 and TM7 to TM11). Galactose is bound at the center of the core and is isolated from external solution by hydrophobic residues.
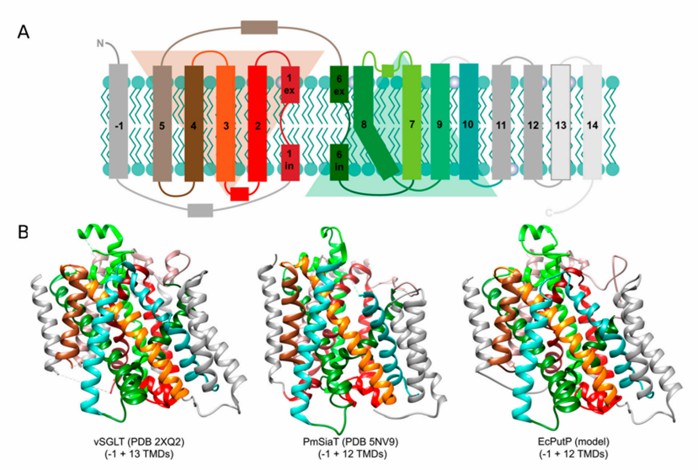 Figure 1. Membrane topology and 3D structures of solute/sodium symporter (SSS) family transporters. (Henriquez T, et al., 2021)
Figure 1. Membrane topology and 3D structures of solute/sodium symporter (SSS) family transporters. (Henriquez T, et al., 2021)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Sodium/Sugar symporter with bound Galactose | Vibrio parahaemolyticus | X-ray diffraction | 2.7 Å | 3DH4 |
| Substrate-bound outward-open state of a Na+-coupled sialic acid symporter | Proteus mirabilis HI4320 | X-ray diffraction | 2.26 Å | 5NVA |
| Substrate-bound outward-open state of a Na+-coupled sialic acid symporter | Proteus mirabilis HI4320 | X-ray diffraction | 1.95 Å | 5NV9 |
| The K294A mutant of vSGLT | Vibrio parahaemolyticus | X-ray diffraction | 2.73 Å | 2XQ2 |
| SGLT2-MAP17 complex bound with substrate AMG in the occluded conformation | Homo sapiens | Cryo-EM single particle analysis | 3.33 Å | 7YNJ |
| SGLT2-MAP17 complex in the apo state in the inward-facing conformation | Homo sapiens | Cryo-EM single particle analysis | 3.48 Å | 7YNK |
| SGLT1-MAP17 complex bound with substrate 4D4FDG in the occluded conformation | Homo sapiens | Cryo-EM single particle analysis | 3.26 Å | 7YNI |
| SGLT1-MAP17 complex bound with LX2761 | Homo sapiens | Cryo-EM single particle analysis | 3.2 Å | 7WMV |
| SMCT1 | Homo sapiens | Cryo-EM single particle analysis | 3.5 Å | 7SL9 |
| SGLT1 | Homo sapiens | Cryo-EM single particle analysis | 3.4 Å | 7SL8 |
| SGLT2-MAP17 complex bound with empagliflozin | Homo sapiens | Cryo-EM single particle analysis | 2.95 Å | 7VSI |
| Bicarbonate transporter complex SbtA-SbtB bound to HCO3- | Synechocystis sp. PCC 6803 substr. Kazusa | X-ray diffraction | 3.2 Å | 7EGL |
| Bicarbonate transporter complex SbtA-SbtB bound to AMP | Synechocystis sp. PCC 6803 substr. Kazusa | Cryo-EM single particle analysis | 2.7 Å | 7EGK |
| A substrate-free glutamate transporter homologue GltTk | Thermococcus kodakarensis KOD1 | X-ray diffraction | 2.7 Å | 5DWY |
| The sodium/iodide symporter (NIS) | Rattus norvegicus | Cryo-EM single particle analysis | 3.3 Å | 7UUY |
| a cysteine-pair mutant (Y113C-P190C) of a bile acid transporter trapped in an outward-facing conformation | Yersinia frederiksenii | X-ray diffraction | 2.861 Å | 6LH1 |
| VcINDY-Na+ in amphipol | Vibrio cholerae | Cryo-EM single particle analysis | 3.16 Å | 6WU3 |
| VcINDY in complex with terephthalate | Vibrio cholerae | X-ray diffraction | 3.92 Å | 6WTX |
| the divalent anion/Na+ symporter | Vibrio cholerae O1 biovar El Tor str. N16961 | X-ray diffraction | 2.78 Å | 5UL9 |
| The divalent anion/Na+ symporter | Vibrio cholerae O1 biovar El Tor str. N16961 | X-ray diffraction | 2.8 Å | 5ULE |
| The sodium/iodide symporter (NIS) in complex with perrhenate and sodium | Rattus norvegicus | Cryo-EM single particle analysis | 3.2 Å | 7UUZ |
| VcINDY-Na-Fab84 in nanodisc | Homo sapiens | Cryo-EM single particle analysis | 3.15 Å | 6WW5 |
| VcINDY-apo | Vibrio cholerae | Cryo-EM single particle analysis | 3.23 Å | 7T9F |
| VcINDY-Na+ | Vibrio cholerae | Cryo-EM single particle analysis | 2.83 Å | 7T9G |
| SVCT1 in an apo state | Mus musculus | Cryo-EM single particle analysis | 3.5 Å | 7YTY |
Table 1. Structural research of the solute sodium symporter (SSS) family.
Creative Biostructure is a renowned organization in the field of structural biology and holds a leading position in providing structural analysis services for membrane proteins. We utilize cutting-edge X-ray crystallography, and cryo-electron microscopy (cryo-EM) techniques to reveal the complex structures of solute sodium symporter (SSS) family members at high resolution. Through our services, we have greatly advanced the understanding of the functional mechanisms of SSS proteins. If you are interested in our services, contact us now to unleash our advanced capabilities and potential to propel you closer to achieving your scientific goals.
References
- Henriquez T, et al. Prokaryotic Solute/Sodium Symporters: Versatile Functions and Mechanisms of a Transporter Family. Int J Mol Sci. 2021. 22(4): 1880.
- Jung H. Towards the molecular mechanism of Na (+)/solute symport in prokaryotes. Biochim Biophys Acta. 2001. 1505(1): 131-143.
- Abramson J, Wright EM. Structure and function of Na (+)-symporters with inverted repeats.Curr Opin Struct Biol. 2009. 19(4): 425-432.