Structural Research of Membrane Trafficking Proteins
Membrane trafficking proteins are responsible for the movement of molecules and structures within the cell membrane and between organelle. Membrane trafficking dysfunction can lead to abnormal conditions in various cells and even the entire body. Therefore, understanding the structure and function of membrane trafficking proteins is crucial for studying the normal physiological and biological activities of cells.
Cryo-EM can capture the inherent flexibility and dynamic characteristics of Membrane trafficking proteins, which makes a particularly important contribution to understanding the molecular mechanisms, drug design, and disease treatment of these proteins. By analyzing multiple images taken from different angles of the same sample, a three-dimensional image of the sample can be reconstructed, and the atomic model of the molecule can be obtained with great certainty.
The movement of signal molecules in and out of cilia is crucial for appropriate signal transduction. The Bardet Biedl Syndrome (BBS) complex is composed of eight proteins called BBSome and plays a crucial role in regulating the withdrawal of certain membrane proteins from cilia.
In 2020, researchers showed that Cryo EM was used to display BBSome itself and the near atomic structure compounded with ARL6GTP at 3.44 Å and 4.00 Å resolutions, respectively, and described the changes in the conformation of BBSome caused by the combination of ARL6GTP. The final experimental results indicate that BBSome-mediated crossing of the ciliary basal transition region is facilitated by the ARF like GTP enzyme ARL6/BBS3. When recruited to the membrane by ARL6GTP, BBSome undergoes a conformational change, which represents its active form. BBSome interacts with G protein coupled receptors (GPCRs) such as Smoothened (SMO) by recognizing a region called amphoteric helix 8 (SMOH8), which must pop out of the inner lobe of the membrane to be recognized by BBSome. This indicates that the interaction between BBSome and its cargo GPCR is a universal principle.
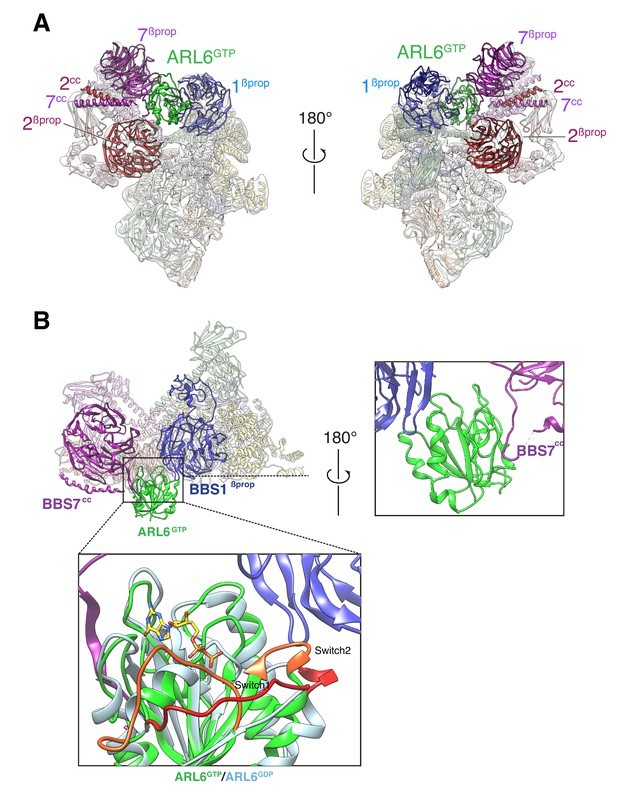 Figure 1. Two views of the cryo-EM map (transparent surface) and the near-atomic model of the BBSome–ARL6GTP complex shown
in ribbon representation. (YANG, et al., 2020)
Figure 1. Two views of the cryo-EM map (transparent surface) and the near-atomic model of the BBSome–ARL6GTP complex shown
in ribbon representation. (YANG, et al., 2020)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| BBSome, apo form | Bos taurus | Cryo-EM single particle analysis | 3.44 Å | 6VNW |
| BBSome-ARL6 complex | Bos taurus | Cryo-EM single particle analysis | 4.00 Å | 6VOA |
Table 1. Structural research of membrane trafficking proteins.
At Creative Biostructure, we can help customers obtain high-resolution structures of membrane proteins. Cryo-electron microscopy (cryo-EM), as a high-resolution biological macromolecule structure analysis technology, has been widely used in the research of membrane proteins. Cryo-EM technology obtains three-dimensional structural information of protein molecules by imaging frozen samples at the atomic level under high vacuum, thereby resolving the spatial structure and functional information of membrane proteins. Cryo-EM also has the inherent flexibility and kinetic ability to capture biological molecules, which is crucial for understanding functionality. By analyzing multiple images of the same sample taken from different angles, a 3D image of the sample can be reconstructed, and then the atomic model of the molecule can be constructed using this 3D image.
Our highly skilled professional team utilizes the latest cryo-EM technology and calculation methods to provide detailed information about the structure and function of membrane proteins. We are committed to making significant contributions to understanding complex biological systems and look forward to working with you.
Reference
- YANG S, et al. Near-atomic structures of the BBSOME reveal the basis for BBSome activation and binding to GPCR cargoes. eLife, 2020, 9.
