Structural Research of Tetraspanins
Tetraspanins are a highly conserved family that play critical roles in a variety of cellular functions, including cell migration, protein transport, maintenance of membrane integrity, and regulation of signal transduction. Members of the tetraspanins family have four transmembrane structural domains, and extracellular loops and bind to a wide range of other chaperone proteins to fulfill the roles. Elucidating the 3D structure of the tetraspanins is an essential step in understanding the molecular basis of their facilitator functions and chaperone specificity.
Advances in structural research of tetraspanins
All tetraspanins have four transmembrane domains, short N-terminal and C-terminal cytoplasmic tails, an intracellular loop, short extracellular loops (SELs), and large extracellular loops (LELs). the C-terminal cytoplasmic tails play a role in sorting and intracellular targeting of tetraspanins in animals. the LELs contain both conserved regions that are involved in homodimerization, and variable regions, which are thought to play a major role in the selection of interaction partners. The variable region is thought to play a major role in interaction partner selection. The variable region contains conserved motifs (animal: GCC, plant: GCCK/RP) and the presence of disulfide bond stabilizing structures. Cryo-electron microscopy data and computational modeling suggest that the TM helices form a tightly linked four-helix bundle.
Crystal structure analysis of human tetraspanins CD9
Researchers obtained the cryo-electron microscopy (cryo-EM) structure of the human tetraspanins CD9 by the lipid cubic phase (LCP) method. The structure shows that CD9 is folded into a quadruple transmembrane helix, with its intracellular ends tightly bundled and its extracellular ends loosely stacked, forming a large central cavity within the intramembrane region. The SEL between TM1 and TM2, and the LEL between TM3 and TM4, extends over this central cavity. LEL is stabilized by disulfide bond pairs between highly conserved cysteine residues.
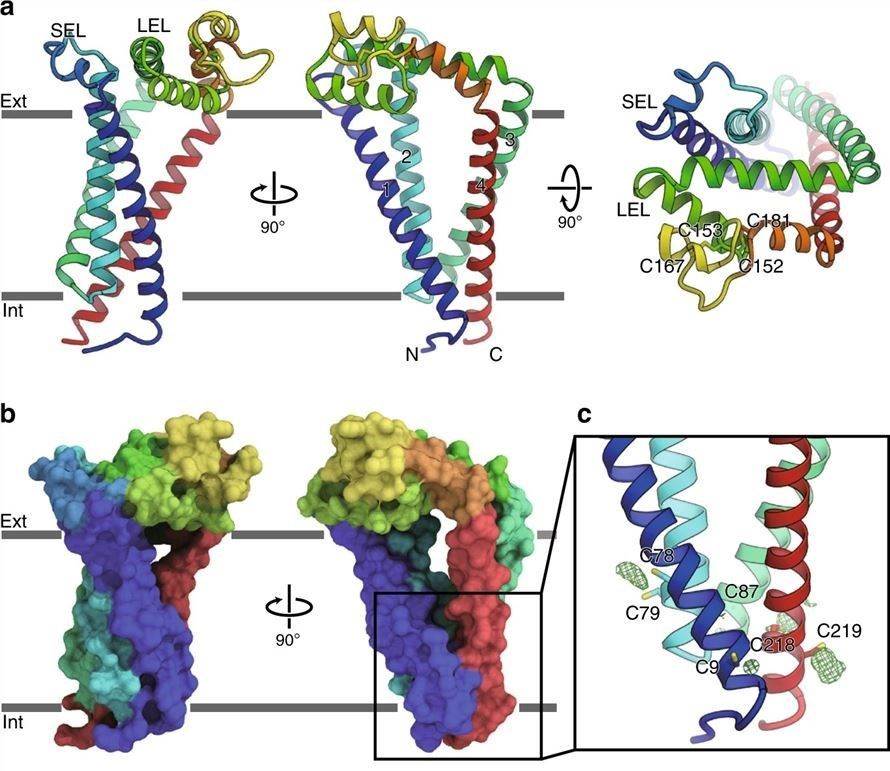 Figure 1. Crystal structure of human tetraspanins CD9. (Umeda R, et al., 2020)
Figure 1. Crystal structure of human tetraspanins CD9. (Umeda R, et al., 2020)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| CD53 | Homo sapiens | X-ray diffraction | 2.9 Å | 6WVG |
| Tetraspanin CD81 | Homo sapiens | X-ray diffraction | 2.955 Å | 5TCX |
| CD81 large extracellular loop (agmCD81-LEL) | Chlorocebus sabaeus | X-ray diffraction | 1.899 Å | 3X0G |
| CD81 large extracellular loop | Saguinus oedipus | X-ray diffraction | 1.8 Å | 7MWS |
| CD81 Large Extracellular Loop. | Homo sapiens | X-ray diffraction | 2.6 Å | 1IV5 |
| CD81 large extracellular loop (mCD81-LEL) | Mus musculus | X-ray diffraction | 1.471 Å | 3X0F |
| CD81 large extracellular loop (hCD81-LEL) | Mus musculus | X-ray diffraction | 1.844 Å | 3X0E |
| CD81LEL (space group P21) | Homo sapiens | X-ray diffraction | 1.28 Å | 5M33 |
| CD81LEL (space group P64) | Homo sapiens | X-ray diffraction | 2.021 Å | 5M3T |
| CD81LEL (space group P32 1 2) | Homo sapiens | X-ray diffraction | 1.961 Å | 5M2C |
| CD81LEL (space group P31) | Homo sapiens | X-ray diffraction | 2.38 Å | 5M3D |
| CD81LEL (space group C2) | Homo sapiens | X-ray diffraction | 3.1 Å | 5M4R |
| CD9 | Homo sapiens | X-ray diffraction | 2.701 Å | 6K4J |
| CD9 large extracellular loop | Homo sapiens | X-ray diffraction | 2 Å | 6RLR |
| Anti-CD9 nanobody 4C8 | Lama glama | X-ray diffraction | 1.7 Å | 6Z1Z |
| EC2 domain of CD9 in complex with nanobody 4E8 | Homo sapiens | X-ray diffraction | 1.33 Å | 6Z1V |
| EC2 domain of CD9 in complex with nanobody 4C8 | Homo sapiens | X-ray diffraction | 2.68 Å | 6Z20 |
| SmTSP2EC2 | Schistosoma mansoni | SOLUTION NMR | / | 2M7Z |
| Tspan15 large extracellular loop (Tspan15 LEL) | Homo sapiens | X-ray diffraction | 2.52 Å | 7RDB |
| Tspan15 large extracellular loop (Tspan15 LEL) in complex with 1C12 Fab | Homo sapiens | X-ray diffraction | 3.6 Å | 7RD5 |
| CD81 extracellular domain, a receptor for hepatitis c virus | Homo sapiens | X-ray diffraction | 1.6 Å | 1G8Q |
| CD81 large extracellular loop in complex with 5A6 Fab | Homo sapiens | X-ray diffraction | 2.4 Å | 6U9S |
| CD81 large extracellular loop in complex with single chain fv fragment 4 | Homo sapiens | X-ray diffraction | 2.82 Å | 6EJG |
| CD81 large extracellular loop in complex with single chain fv fragment 5 | Homo sapiens | X-ray diffraction | 2.15 Å | 6EJM |
| CD81 large extracellular loop in complex with single chain fv fragment 10 | Homo sapiens | X-ray diffraction | 2.65 Å | 6EK2 |
| CD19-CD81 co-receptor complex bound to coltuximab Fab fragment | Homo sapiens | Cryo-EM single particle analysis | 3.8 Å | 7JIC |
| ADAM10-Tspan15 complex bound to 11G2 vFab | Homo sapiens | Cryo-EM single particle analysis | 3.3 Å | 8ESV |
Table 1. Structural research of the tetraspanins.
Creative Biostructure, as an expert in structure elucidation, specializes in providing high-quality structural biology solutions to support drug discovery, protein engineering, and basic research. Our experienced scientists are well-versed in a variety of techniques such as X-ray crystallography, cryo-electron microscopy (cryo-EM), and NMR spectroscopy. Whether you require a full range of structural biology support services or project-specific expertise, we can satisfy your specific requirements.
In addition to tetraspanins, we can provide structural analysis services for a wide range of biomolecules. We are committed to providing timely and accurate results and maintaining open communication throughout the project. Please feel free to contact us with any questions or to discuss your project in more detail.
References
- Umeda R, et al. Structural insights into tetraspanin CD9 function. Nat Commun. 2020. 11(1): 1606.
- Reimann R, et al. TETRASPANINs in Plants. Front Plant Sci. 2017. 8: 545.
- Susa KJ, et al. Tetraspanins: structure, dynamics, and principles of partner-protein recognition. Trends Cell Biol. Published online September 30, 2023.
