Chromatin Structure
Chromatin is a complex and dynamic substance within the nucleus of eukaryotic cells that plays a critical role in regulating gene expression, DNA replication, and repair. It is composed of deoxyribonucleic acid (DNA) and associated proteins, primarily histones, which together form a highly organized and flexible structure. An understanding of chromatin structure is fundamental to the comprehension of genetic information regulation, with significant implications for health, disease, and the development of novel therapeutic strategies. In addition to the structure of chromatin, other significant structures within the nucleus include the nuclear pore complex and the nuclear skeleton.
What is Chromatin?
Chromatin is defined as the substance of chromosomes, consisting of DNA wrapped around histone proteins. Its primary function is to package DNA into a more compact and denser configuration, thereby enabling its accommodation within the nucleus of eukaryotic cells. In addition to its role in packaging DNA, chromatin plays a crucial part in regulating genetic processes. It facilitates the organization of the genome and controls gene expression by modifying the accessibility of DNA to transcriptional machinery.
Chromatin exists in two primary forms: euchromatin and heterochromatin. Euchromatin is less condensed and is typically associated with active gene transcription. In contrast, heterochromatin is more compact and typically associated with transcriptional repression and structural roles, such as centromeres and telomeres.
Organization of Chromosomes from DNA
The organization of chromosomes commences with the DNA double helix, which is subsequently condensed into a higher-order structure through interactions with histone proteins. The fundamental unit of chromatin is the nucleosome, which is formed by the winding of DNA around a core structure. This structure is fundamental to an understanding of the processes by which chromatin organizes and compacts DNA.
DNA-Histone Interactions: DNA wraps around histone octamers, which consist of two copies of each of the histones H2A, H2B, H3, and H4. This wrapping creates a "bead-on-a-string" structure, with each nucleosome acting as a bead. The DNA in this structure is approximately 147 base pairs long and is wrapped around the histone core.
Nucleosome and Linker DNA: Between each nucleosome bead is a segment of linker DNA that connects successive nucleosomes and can vary in length. Histone H1 binds to this linker DNA and further stabilizes the nucleosome structure, contributing to the formation of the 30 nm fiber, a more compact structure of chromatin.
Higher-Order Chromatin Organization: The 30 nm fiber folds into loops and domains that organize into more complex structures, ultimately forming the chromosomes visible during cell division. This organization is critical for maintaining genomic stability and regulating gene expression.
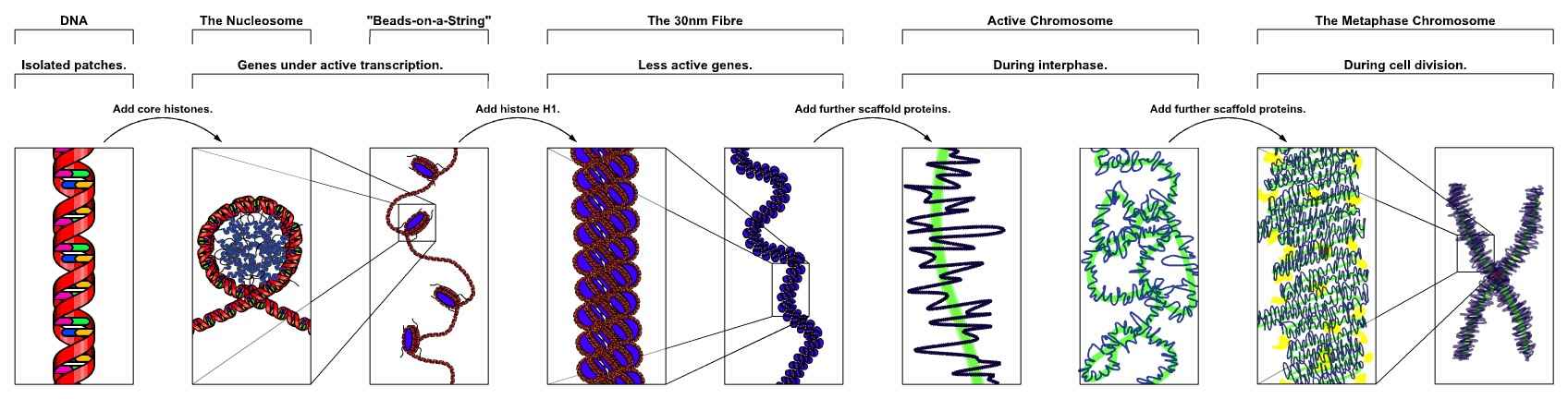 Figure 1. The major structures in DNA compaction: DNA, the nucleosome, the 11 nm beads on a string chromatin fiber and the metaphase chromosome.
Figure 1. The major structures in DNA compaction: DNA, the nucleosome, the 11 nm beads on a string chromatin fiber and the metaphase chromosome.
Nucleosome: The Fundamental Unit of Chromatin
The nucleosome is the basic unit of chromatin structure, consisting of DNA wrapped around a core of histone proteins. Each nucleosome core is composed of an octamer of histones, providing a stable platform for DNA wrapping. The interaction between histones and DNA is primarily through electrostatic forces and hydrogen bonds, allowing for a compact yet flexible structure.
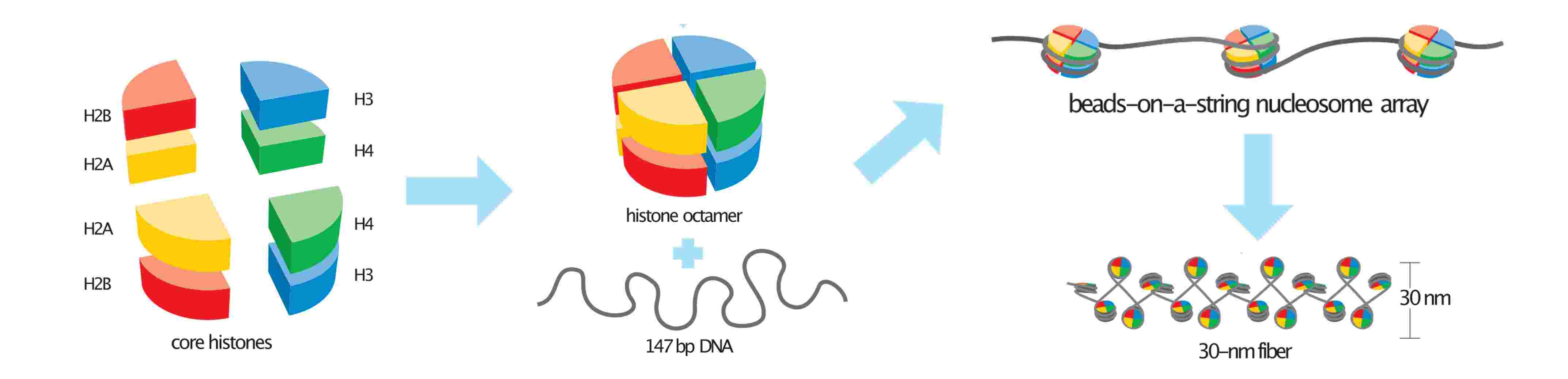 Figure 2. Basic units of chromatin structure.
Figure 2. Basic units of chromatin structure.
There are a variety of modifications in the organization of the nucleosome that can affect chromatin structure and function including histone modifications and histone variants. Histones undergo various post-translational modifications, including acetylation, methylation, and phosphorylation. These modifications can influence chromatin structure and function by altering histone-DNA interactions and recruiting other regulatory proteins. In addition to canonical histones, there are histone variants that can replace standard histones in nucleosomes. These variants often play specialized roles in chromatin function, such as regulating gene expression or DNA repair.
Dynamic Chromatin Structure During the Cell Cycle
Chromatin is highly dynamic in proliferating cells. Two major events occur during the cell cycle that allow for global chromatin remodeling. First, the incorporation of new histones onto nascent DNA occurs during S phase and creates a requirement for the reestablishment of histone PTMs. Second, many chromatin remodeling complexes and transcriptional complexes are dissociated from chromatin during mitosis, and nuclear architecture, including chromatin domains or nuclear interior/periphery associations, is disrupted.
Specifically, cells in G1 phase exhibit subnuclear regions associated with nuclear pores and the nuclear lamina. The pre-replication complex (pre-RCs), a group of proteins that initiate DNA replication at specific origins, tends to form on more accessible chromatin. During S phase, histones are transcribed and synthesized, DNA is replicated, and new nucleosomes are assembled with recycled ones to form new chromatin. Enzymes that add or remove post-translational modifications (PTMs) also associate with this newly formed chromatin. In G2, nucleosomes become fully mature and histone production is halted. During mitosis, chromosomes condense and many transcription factors and proteins that bind to chromatin are released. The nuclear envelope disassembles, disrupting the domains associated with the nuclear lamina.
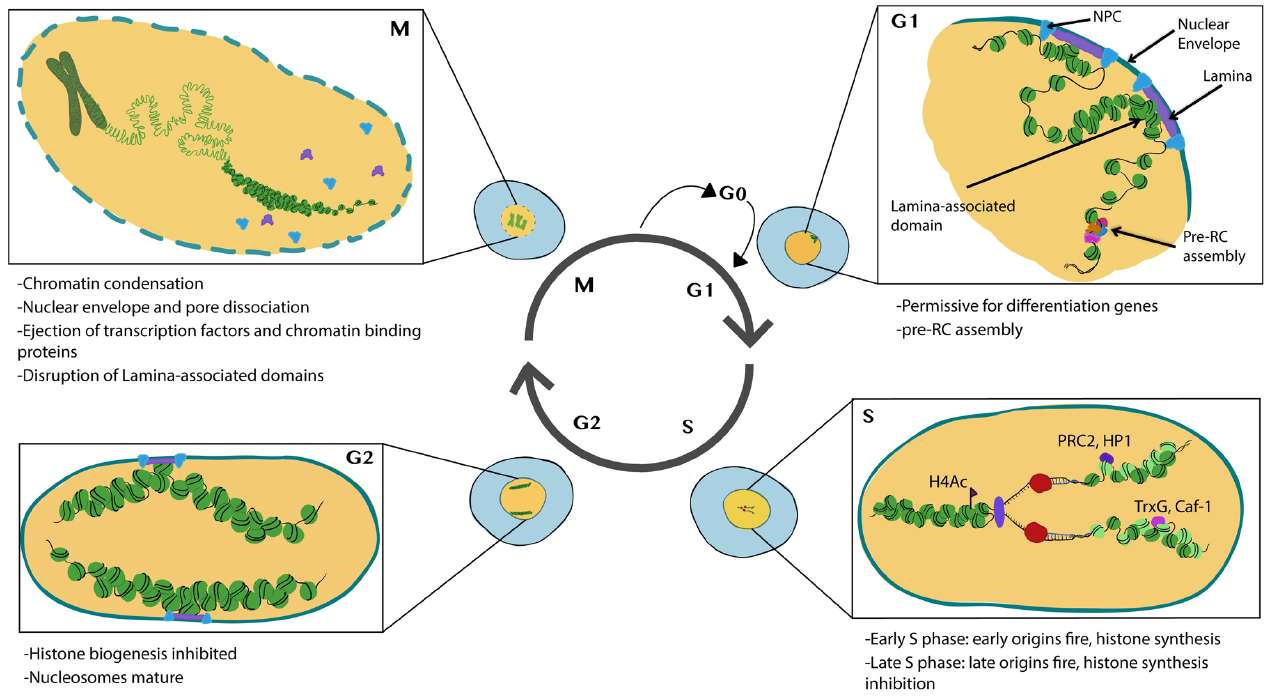 Figure 3. Major features of chromatin and nuclear changes during the cell cycle. New assembled nucleosomes: light green; recycled nucleosomes: dark green (Ma, Kanakousaki, and Buttitta, 2015).
Figure 3. Major features of chromatin and nuclear changes during the cell cycle. New assembled nucleosomes: light green; recycled nucleosomes: dark green (Ma, Kanakousaki, and Buttitta, 2015).
Methods for Analyzing Chromatin Structure
Understanding chromatin structure and dynamics requires a variety of analytical techniques. Each method provides unique insights into different aspects of chromatin organization, modification, and function.
- Chromatin Immunoprecipitation (ChIP): This antibody-based technology is employed to selectively enrich specific DNA-binding proteins along with their DNA targets. One objective is to ascertain whether particular proteins are associated with specific gene regions. For instance, the binding of a transcription factor to promoters, enhancers, repressors, or silencers can be investigated. Another significant field of research is the examination of the distribution of different histone modifications, which constitute a basis of epigenetic gene regulation.
- Chromatin Accessibility Assays: Techniques such as ATAC-seq (Assay for Transposase-Accessible Chromatin using sequencing) and DNase-seq are employed to assess chromatin accessibility. These methods identify genomic regions that are open and accessible to regulatory proteins, thereby providing insights into transcriptional regulation and chromatin structure.
- DNA Foot Printing: This method is aimed at identifying protein-bound DNA. It uses labeling and fragmentation coupled to gel electrophoresis to identify areas of the genome that have been bound by proteins. Techniques like DNA foot printing help elucidate which proteins bind to these associated regions of DNA and unravel the complexities of transcriptional control.
- Chromosome Conformation Capture (3C) Techniques: This technology determines the spatial organization of chromatin in the nucleus by inferring genomic locations that physically interact. Hi-C is the first 3C derivative technology to be truly genome-wide. Hi-C is a technique for studying the 3D organization of the genome. By capturing chromatin interactions across the entire genome, Hi-C provides a comprehensive view of chromatin folding and the spatial organization of chromosomes within the nucleus.
- Fluorescence In Situ Hybridization (FISH): FISH is used to visualize specific DNA sequences within chromosomes. By using fluorescent probes, researchers can analyze chromosome structure, identify chromosomal abnormalities, and study gene localization.
- Microscopy Techniques: Advanced microscopy techniques, such as super-resolution microscopy, allow the visualization of chromatin at high resolution. These techniques can reveal the spatial organization of chromatin within the nucleus and provide insight into chromatin dynamics during various cellular processes.
- Mass Spectrometry: Mass spectrometry is utilized to analyze histone modifications and identify chromatin-associated proteins. This technique enables the profiling of post-translational modifications and facilitates the elucidation of the functional implications of these modifications.
In conclusion, chromatin represents a fundamental component of eukaryotic cells, playing an intricate role in the regulation of gene expression, DNA replication, and repair. A comprehensive understanding of its structure, from the fundamental nucleosome unit to higher-order organization, is essential for elucidating its function in cellular processes. The dynamic alterations in chromatin during the cell cycle serve to illustrate its flexibility and adaptability.
At Creative Biostructure, we provide comprehensive analyses of chromatin organization, including nucleosome positioning, histone modifications, and chromatin accessibility. Our cutting-edge technologies and expert team are dedicated to delivering precise, actionable insights that drive your discoveries forward. Whether you're studying gene regulation, DNA repair mechanisms, or cellular dynamics, our tailored solutions ensure you have the data you need to excel. Contact us today to learn how our advanced chromatin structure testing can enhance your research and support your biopharmaceutical innovations.
References
- Molecular Biology of the Cell, 6th Edition.
- Ma, Y., Kanakousaki, K., & Buttitta, L. (2015). How the cell cycle impacts chromatin architecture and influences cell fate. Frontiers in Genetics, 6.
- Minnoye, L., Marinov, G. K., Krausgruber, T., Pan, L., Marand, A. P., Secchia, S., Greenleaf, W. J., Furlong, E. E. M., Zhao, K., Schmitz, R. J., Bock, C., & Aerts, S. (2021). Chromatin accessibility profiling methods. Nature Reviews Methods Primers, 1(1), 10.