Structural Research of Guanosine Triphosphatases
How cell growth is regulated is one of the most fundamental issues in life science. This process is mainly controlled by the TOR (Target of Rapamycin) signaling pathway. In 2008, two research groups independently discovered that Rag GTPases can mediate amino acid activation of mTORC1, filling the gap in signal transmission between amino acids and mTORC1.
The Rag GTPases subfamily belongs to the Ras small G protein family. There are four members of Rag GTPase in the human genome, namely RagA, RagB, RagC, and RagD. Among them, RagA and RagB are highly homologous, while RagC and RagD are highly homologous. RagA or RagB can form specialized dimer complexes with RagC or RagD. It is worth mentioning that Rag GTPases is the only small G protein that can form dimers in the Ras small G protein family as we know it.
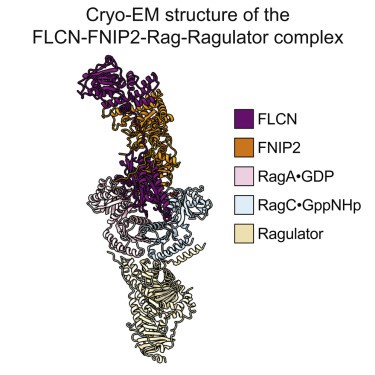 Figure 1. Cryo-EM structure of the FLCN-FNIP2-Rag-Ragulator complex. (SHEN K, et al., 2019)
Figure 1. Cryo-EM structure of the FLCN-FNIP2-Rag-Ragulator complex. (SHEN K, et al., 2019)
mTORCl (mTOR complex1) responds to nutrients by controlling synthetic and catabolic processes through the Rag GTP enzyme heterodimer, which is regulated by multiple upstream protein complexes. The structure of FLCN-FNIP2 in complexes with Rag GTPases and Ragulator has been determined using a cryo-electron microscope. The research results have helped researchers understand the mechanism by which FLCN-FNIP2 activates Rag GTP enzyme heterodimers.
The FLCN-FNIP2-Rag-Ragulator complex adopts a slender conformation with a size of 220 × 100 × 60 Å. The Rag GTPase heterodimer is located at the center. Their CRD interacts with Ragulator, similar to what was observed in the previously analyzed structure FLCN-FNIP2 binds to Rag GTPase through its nucleotide-binding domain (NBD). Although FLCN-FNIP2 is a specific GAP of RagC, it enters the space between Rag subunits and directly contacts the NBD of RagA and RagC, indicating that it functions through a unique molecular mechanism.
This structural model provides insight into the heterodimerization mechanism between FLCN and FNIP2. We have identified two contact surfaces between FLCN and FNIP2, each formed by heterodimerization of its corresponding structural domain. Firstly, FLCN and FNIP2 each contribute a Longin domain to form heterodimers, which directly enter the space between the NBD of RagA and RagC. This heterodimerization is mediated by a continuous ten-chain β folding as an intermediate platform, sandwiched between 6 α helices.
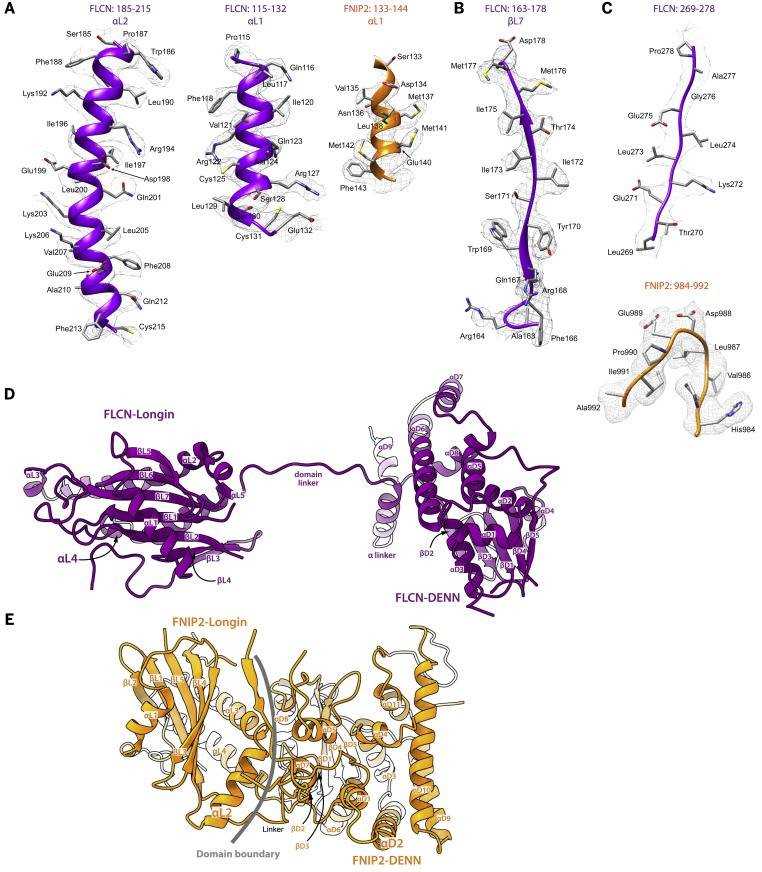 Figure 2. Structural Model for FLCN-FNIP2. (SHEN K, et al., 2019)
Figure 2. Structural Model for FLCN-FNIP2. (SHEN K, et al., 2019)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Lysosomal Folliculin Complex (FLCN-FNIP2-RagA-RagC-Ragulator) | Homo sapiens | Cryo-Electron Microscopy | 3.60 Å | 6NZD |
| FLCN-FNIP2-Rag-Ragulator complex | Homo sapiens | Cryo-Electron Microscopy | 3.31 Å | 6ULG |
Table 1. Structural Research of Guanosine Triphosphatases.
Do you need high-resolution membrane protein structure analysis services? Creative Biostructure provides customers with membrane protein analysis services based on cryo-electron microscopy technology. Cryo-electron microscopy technology rapidly freezes samples in liquid nitrogen to preserve their structure, and then uses electron microscopy to capture high-resolution images of these samples, making it a powerful imaging technique. If you are interested in learning more about our protein structural analysis services, please contact us for more information.
References
- LAWRENCE R E, et al. Structural mechanism of a rag GTPase activation checkpoint by the lysosomal Folliculin Complex. Science, 2019, 366(6468): 971–977.
- SHEN K, et al. Cryo-EM structure of the human FLCN-FNIP2-rag-Ragulator Complex. Cell, 2019, 179(6).