Structural Research of Hepeviridae
The Hepeviridae comprise small, envelope-less positive-sense RNA viruses. Members of the family are divided into two genera, Piscihepevirus, which infects fish, and Orthohepevirus, which infects mammals and birds. Of these, the most widely researched is hepatitis E virus (HEV), one of the common causes of acute viral hepatitis. HEV are zoonotic pathogens with a wide range of hosts. In recent years, tremendous progress has been made in the biological and structural characterization of HEV. Determination of the high-resolution three-dimensional structure of HEV virus-like particles (VLPs) can help to understand morphogenesis and pathogenesis, which is essential for the development of universal strategies for preventing HEV infections and the discovery of prophylactic vaccines.
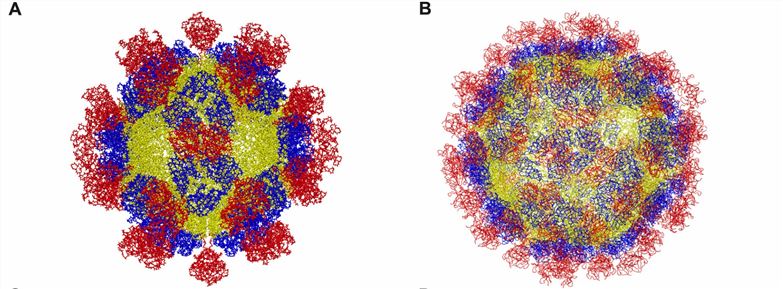 Figure 1. Structure of hepatitis E virus. (A) T = 1. (B) T = 3. (Wang B, Meng XJ., 2021)
Figure 1. Structure of hepatitis E virus. (A) T = 1. (B) T = 3. (Wang B, Meng XJ., 2021)
Genome Structure of Hepeviridae Members
The genomes of Hepeviridae members are ~6.4 to 7.2kb single-stranded positive-sense RNAs with three major open reading frames (ORF1, ORF2, and ORF3). The ORF1 encodes a viral nonstructural protein with an upstream RNA structural element that binds to capsid protein (CP). ORF2 encodes the structural CP, and the ORF2 and ORF3 proteins are expressed by subgenomic mRNAs. The region connecting ORF1 and ORF3 contains elements that control the expression of subgenomic biallelic mRNAs. ORF3 overlaps with the 5' coding sequence of ORF2, and neither ORF3 nor ORF2 overlaps with ORF1. A cis-active element found within the 3' UTR overlapping with the carboxy-terminal sequence of ORF2 is involved in viral RNA replication.
Features and Structure of Hepeviridae Viral Particles
For example, HEV virus particles are characterized by the expression of truncated CPs in insect cells, leading to the self-assembly and generation of two types of virus-like particles (VLPs), T = 1 and natural virus particle-sized T = 3. The cryo-electron microscopy (cryo-EM) structures of T = 3 VLPs have been resolved. The structures show that the viral particles consist of 180 CP copies and include three functional domains, S (shell), M (middle), and P (protruding). The S domain forms an icosahedral shell and is shown to be a jelly roll fold common to small RNA viruses. The P domain is hypothesized to be a binding site for the receptor and is further divided into P1 with a triple prominence and P2 with a double cusp. Both P regions have a β-barrel fold and potential polysaccharide binding sites. The M domain is strongly related to the S and P structural domains. The M domain interacts strongly with the S and P domains and may contribute to the stability of the virus particle.
As a leader in viral structure elucidation, Creative Biostructure offers high-quality, comprehensive virus-like particles (VLPs) products that play an essential role in viral structure research. Whether you are researching the molecular mechanisms underlying virus entry or antiviral drug and vaccine discovery, our products will accelerate your research efforts.
| Cat No. | Product Name | Virus Name | Source | Composition |
| CBS-V078 | HEV VLP (Capsid Protein) | Hepatitis E virus | Mammalian cell recombinant | Capsid Protein |
| CBS-V610 | HEV VLP (p239 Proteins) | Hepatitis E virus | E. coli recombinant | p239 |
| CBS-V614 | Hepatitis E virus VLP (ORF2 (112-660aa) Proteins) | Hepatitis E virus | Insect cell recombinant | ORF2 (112-660aa) |
| Explore All Hepeviridae Virus-like Particle Products | ||||
Creative Biostructure has cutting-edge structural biology platforms for mainstream technologies. Our virus-like particles (VLPs) products provide researchers with controlled and reproducible systems to study the overall structure of viruses. Through advanced cryo-electron microscopy (cryo-EM), our scientists can analyze the size, shape, and surface features of virus particles or virus-like particles at high resolution, helping clients understand the assembly, organization, and architecture of viral components.
In addition, we provide clients with tailor-made VLP construction services. If the VLPs you need are not on the product list, please feel free to contact us.
References
- Wang B, Meng XJ. Structural and molecular biology of hepatitis E virus. Comput Struct Biotechnol J. 2021. 19: 1907-1916.
- Kenney SP, Meng XJ. Hepatitis E Virus Genome Structure and Replication Strategy. Cold Spring Harb Perspect Med. 2019. 9(1): a031724.
- Purdy MA, et al. ICTV Virus Taxonomy Profile: Hepeviridae 2022. J Gen Virol. 2022. 103(9): 10.1099/jgv.0.001778.
- Cancela F, et al. Structural aspects of hepatitis E virus. Arch Virol. 2022. 167(12): 2457-2481.
