Flag-tagged Proteins
A cornerstone of modern molecular biology, flag-tagging enables the precise detection, purification and functional analysis of proteins. Introduced in the late 20th century, it has rapidly become an indispensable tool for researchers unraveling the complexities of cellular and molecular processes. At its core, Flag-tagging uses a small peptide sequence, known as a FLAG tag, to confer unique properties to the protein of interest.
Creative Biostructure provides a range of protein tagging options, including GST-tagging, His-tagging, MBP-tagging, and Flag-tagging, tailored to meet your custom requirements.
Principles of Flag-tagging
Flag-tagging involves the fusion of a short, hydrophilic peptide, typically 8-16 amino acids in length, to a target protein. The FLAG tag sequence is usually DYKDDDDK, which is characterized by high hydrophilicity and minimal interference with protein function. The tag can be attached to the N-terminus or C-terminus of the protein of interest, depending on experimental requirements.
The efficacy of the FLAG tag lies in its ability to bind monoclonal antibodies with high specificity. Antibodies such as anti-FLAG M2 readily recognize the epitope, facilitating downstream applications. The small size of the tag minimizes the risk of perturbing protein structure or function, a critical factor when studying native protein behavior. In addition, the sequence contains several negatively charged aspartate residues that enhance its solubility in aqueous environments.
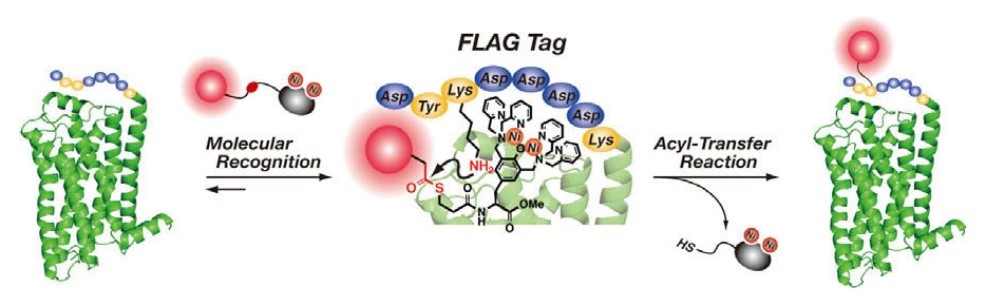 Figure 1. Selective covalent labeling of a FLAG tag fused protein via an acyl-transfer reaction. (Nonaka et al., 2009)
Figure 1. Selective covalent labeling of a FLAG tag fused protein via an acyl-transfer reaction. (Nonaka et al., 2009)
Why Flag-tagging?
The utility of Flag-tagging stems from its versatility and precision. Proteins, by their nature, are often challenging to isolate and study due to their heterogeneity and complex interactions within the cell. Flag-tagging simplifies this process by providing a universal "handle" for protein purification and detection.
Flag-tagging boasts several key advantages, setting it apart from alternative tagging methods such as His-tags or GFP fusion:
- Small Size: Unlike bulky fluorescent proteins, the FLAG-tag minimally disrupts protein folding and function.
- High Affinity and Specificity: The FLAG-tag's robust interaction with its antibody ensures reliable detection and purification.
- Wide Applicability: Its compatibility with multiple systems and techniques enhances its utility.
- Ease of Detection: Anti-FLAG antibodies are commercially available and highly effective, simplifying experimental workflows.
- Reproducibility: Standardized protocols and reagents contribute to consistent, reproducible results.
Applications of Flag-tagged Proteins
Protein Purification
Flag-tagged proteins are revolutionizing the purification process. By exploiting the high-affinity interaction between the FLAG-tag and anti-FLAG antibodies, researchers can obtain nearly homogeneous protein samples. Affinity chromatography using anti-FLAG antibody-conjugated resins enables one-step purification while preserving protein integrity and activity. This is especially important for proteins that are prone to denaturation or degradation during traditional purification methods.
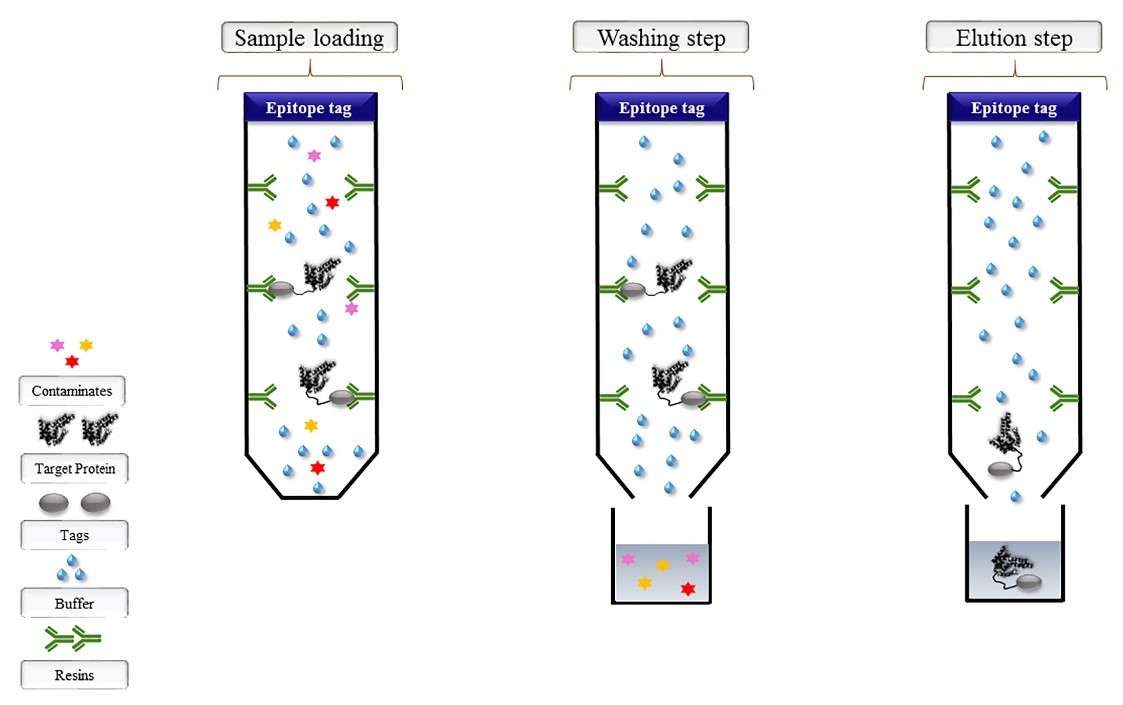 Figure 2. Epitope tag affinity chromatography. (Mahmoudi Gomari et al., 2020)
Figure 2. Epitope tag affinity chromatography. (Mahmoudi Gomari et al., 2020)
Protein Localization
Understanding where a protein is located in the cellular environment provides insight into its function. Flag-tagged proteins, visualized with fluorescence-labeled anti-FLAG antibodies, allow precise mapping of subcellular localization. For example, mitochondrial or nuclear targeting signals fused to FLAG-tags have elucidated protein dynamics under various physiological conditions.
Protein-Protein Interaction Studies
Flag-tagging simplifies the elucidation of protein interaction networks. Co-immunoprecipitation (co-IP) assays leverage Flag-tagged bait proteins to pull down their binding partners. When combined with mass spectrometry, this approach identifies interacting proteins with remarkable accuracy, revealing intricate signaling pathways.
Functional Studies and High-Throughput Screening
In functional genomics and proteomics, Flag-tagged proteins accelerate the screening of gene and protein function. Libraries of Flag-tagged constructs allow systematic testing of thousands of proteins in parallel, providing insight into cellular processes such as gene expression, enzyme activity, and signal transduction.
Recent Innovations and Future Directions
The field of protein tagging continues to evolve, with innovations enhancing the efficiency and scope of FLAG-tag applications.
Dual-Tagging Strategies
Dual-tagging, wherein FLAG-tags are combined with other epitopes (e.g., His-tags or HA-tags), allows sequential or simultaneous purification and analysis. This strategy provides added flexibility for complex experimental designs, such as co-localization studies.
Integration with Synthetic Biology
In synthetic biology, FLAG-tags are increasingly used to engineer multifunctional proteins with tailored properties. Applications range from designer enzymes to synthetic signaling pathways, expanding the horizons of protein engineering.
Proteomics Applications
FLAG-tags have become an integral part of proteomics workflows. For example, proximity labeling techniques such as BioID use Flag-tagged enzymes to biotinylate nearby proteins, enabling the identification of spatially localized interactomes.
Advanced Detection Techniques
Emerging technologies such as single-molecule fluorescence microscopy and cryo-electron microscopy benefit from Flag-tagging. The tag's small size and ease of antibody conjugation make it an ideal choice for high-resolution imaging.
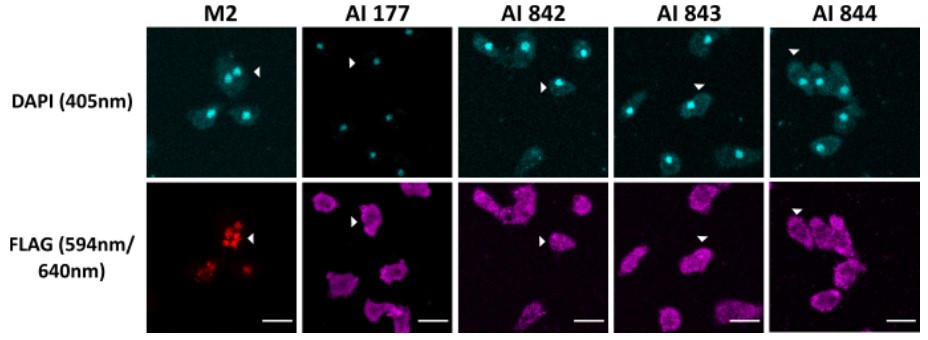 Figure 3. Immunofluorescence microscopy of cells expressing a Flag-tagged protein. The commercial M2 anti-FLAG (red) was used as a positive control. Negative controls of the secondary antibodies did not reveal any signals. AI177, AI842, AI843 and AI844 recombinant antibodies (pink). In cells expressing LmpA-FLAG, the protein is accumulated in a postlysosomal compartment. The arrowheads point to the cells expressing the FLAG-tagged proteins. Representative confocal images of Ax2(Ka) cells stained with DAPI (blue) and anti-FLAG. Scale bar: 10µm. (Mottet and Soldati, 2020)
Figure 3. Immunofluorescence microscopy of cells expressing a Flag-tagged protein. The commercial M2 anti-FLAG (red) was used as a positive control. Negative controls of the secondary antibodies did not reveal any signals. AI177, AI842, AI843 and AI844 recombinant antibodies (pink). In cells expressing LmpA-FLAG, the protein is accumulated in a postlysosomal compartment. The arrowheads point to the cells expressing the FLAG-tagged proteins. Representative confocal images of Ax2(Ka) cells stained with DAPI (blue) and anti-FLAG. Scale bar: 10µm. (Mottet and Soldati, 2020)
Flag-tagging has transformed the study of proteins, bridging gaps in our understanding of cellular and molecular biology. Its precision, versatility, and reproducibility make it an invaluable tool in research, from protein purification to functional genomics.
Creative Biostructure is excited to support your Flag-tagged protein research, from expression vector construction to high-yield recombinant protein production. Contact us today for more information.
References
- Nonaka H, Fujishima S hei, Uchinomiya S hei, Ojida A, Hamachi I. FLAG-tag selective covalent protein labeling via a binding-induced acyl-transfer reaction. Bioorganic & Medicinal Chemistry Letters. 2009;19(23):6696-6699.
- Mahmoudi Gomari M, Saraygord-Afshari N, Farsimadan M, Rostami N, Aghamiri S, Farajollahi MM. Opportunities and challenges of the tag-assisted protein purification techniques: Applications in the pharmaceutical industry. Biotechnology Advances. 2020;45:107653.
- Mottet M, Soldati T. AI842, AI843, AI844 and AI177 antibodies do not recognize a FLAG-tagged protein by immunofluorescence in D. discoideum cells. Antib Rep. 2020;3(2):e126.