Multi-omics Technologies in Exosome Analysis
Mechanistic studies of exosomes involve exploring scientific questions at multiple levels and ways. It starts with confirming the biological function of exosomes and then analyzes how exosomes function. While the former is mainly a phenotypic analysis, the latter includes changes at the molecular level that are affected by exosomes and what are the key elements in exosomes that carry out their functions. In recent years, histologic techniques have provided an efficient strategy for studying how exosomes function.
Biogenesis Pathway and Mechanism of Action of Exosomes
Exosomes are small vesicles about 30~150nm in diameter secreted by cells. Exosomes are rich in proteins, lipids, DNA, and various RNAs (miRNA, lncRNA, circRNA, mRNA, etc.). Exosomes from blood, urine, amniotic fluid, breast milk, and ascites of animal fluids usually contain specific RNAs and proteins that reflect the health or disease state of the organism. Exosomes are generally targeted to bind to receptor cells via surface membrane proteins. The contents of the exosome are delivered to the recipient cell through receptor-ligand interaction, endocytosis, or fusion with the membrane of the target cell, thereby causing corresponding functional changes. This cell-to-cell delivery activity plays an important regulatory role in a wide range of human health and disease domains, including development, immunity, tissue homeostasis, and tumor progression.
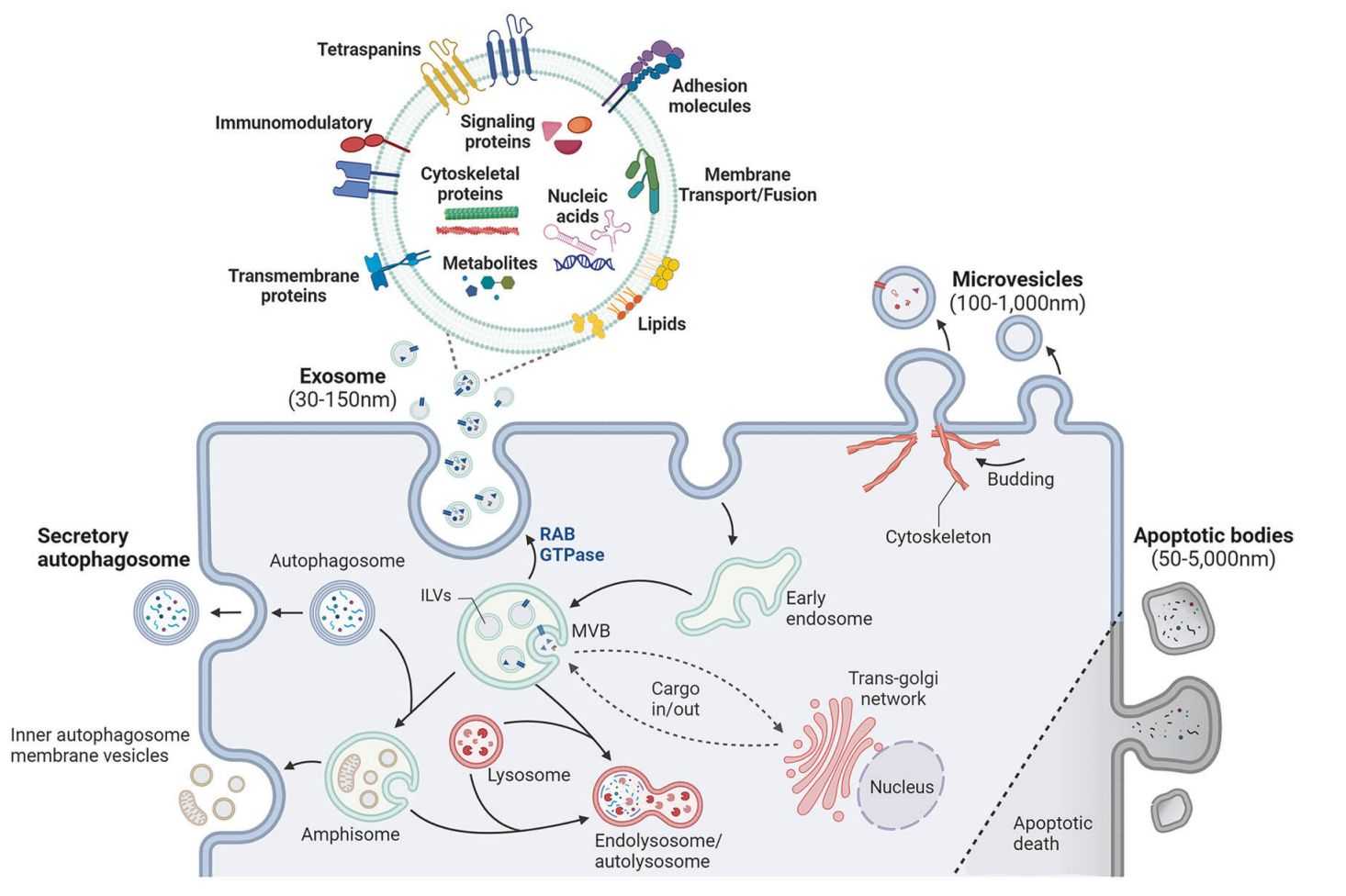 Figure 1. Mechanisms and components of exosome biosynthesis. (Lee YJ, et al., 2024)
Figure 1. Mechanisms and components of exosome biosynthesis. (Lee YJ, et al., 2024)
Significance of Using Multi-omics Techniques to Study Exosomes
- Comprehensive analysis of exosomal cargo
Multi-omics approaches, such as proteomics, transcriptomics, and metabolomics, allow for detailed characterization of the various biomolecules (proteins, RNAs, lipids, and metabolites) present in exosomes, revealing the diverse functions and potential therapeutic applications of exosomes.
- Characterization of exosome biomarkers
Multi-omics analysis helps to identify unique exosome features or biomarkers associated with specific diseases or physiological conditions for early disease detection, monitoring disease progression, and assessing therapeutic response.
- Elucidation of exosome biosynthesis and transport
Multi-omics technologies can provide insights into the cellular pathways and molecular mechanisms involved in exosome biosynthesis, loading, and transport, and inform the development of strategies for designing exosomes for targeted drug delivery.
- Optimization of exosome drug loading and targeting
Multi-omics data can help determine the optimal biomolecular composition and surface properties of exosomes for efficient drug loading and targeted delivery to specific cell types or tissues, which is important for the design and manufacture of exosome-based drug delivery systems with enhanced therapeutic effects.
Exosomes as Biomarkers for Disease Diagnosis
Exosomes are present in various cell types, including tumor cells. They can be isolated from the body fluids of a wide range of organisms, and their primary role is to participate in intercellular substance transport and information transfer. Exosomes carry a large number of specific proteins, nucleic acids, signaling molecules, lipids, etc. They can reflect abnormalities at the cellular level and are closely associated with the onset and progression of many diseases. Therefore, exosomes can be used as biomarkers for disease diagnosis. As an emerging in vitro diagnostic technology, exosomes have great prospects for application in the screening and diagnosis of tumors and other chronic diseases.
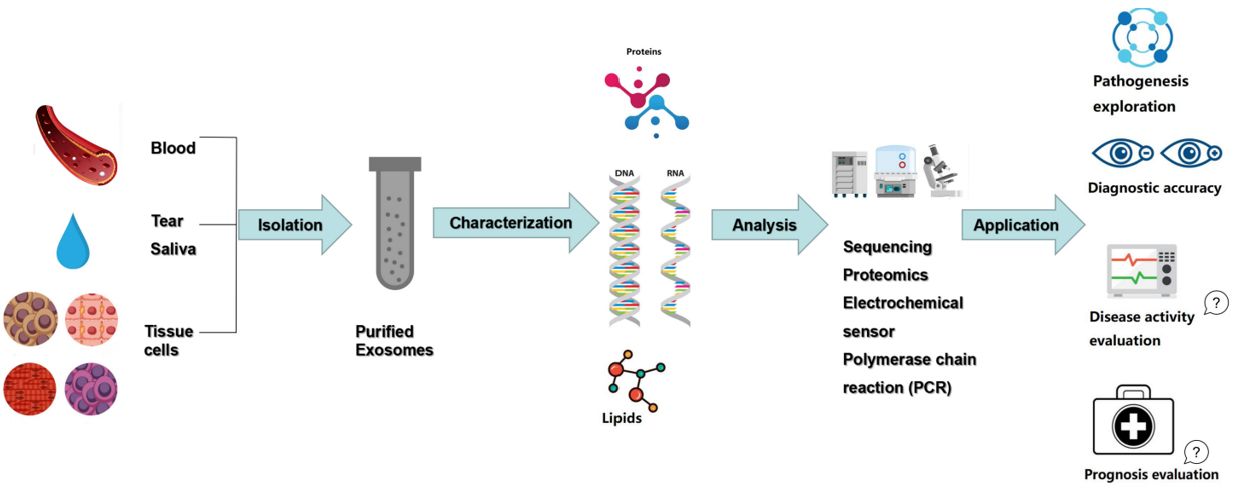 Figure 2. Exosomes as novel diagnostic biomarkers. (Huang Y, et al., 2020)
Figure 2. Exosomes as novel diagnostic biomarkers. (Huang Y, et al., 2020)
Proteomic Analysis of Exosomes
Considering the limitations of exosome isolation techniques, mass spectrometry-based proteomics approaches are increasingly being used for exosome studies. In addition to a conserved set of common proteins critical for biosynthesis, structure, and transport, exosomes contain proteins specific to biological sources. The identification and characterization of these proteins provide important information about the molecular mechanisms involved in cargo sorting and transport, as well as clues about target cells. High-throughput proteomic analysis of exosomes will provide information about the differential expression of proteins and their post-translational modifications. Comparative understanding of exosomal proteomes from different clinical conditions may be valuable for the development of potential biomarkers for early diagnosis, disease progression, prognosis, and therapeutic response.
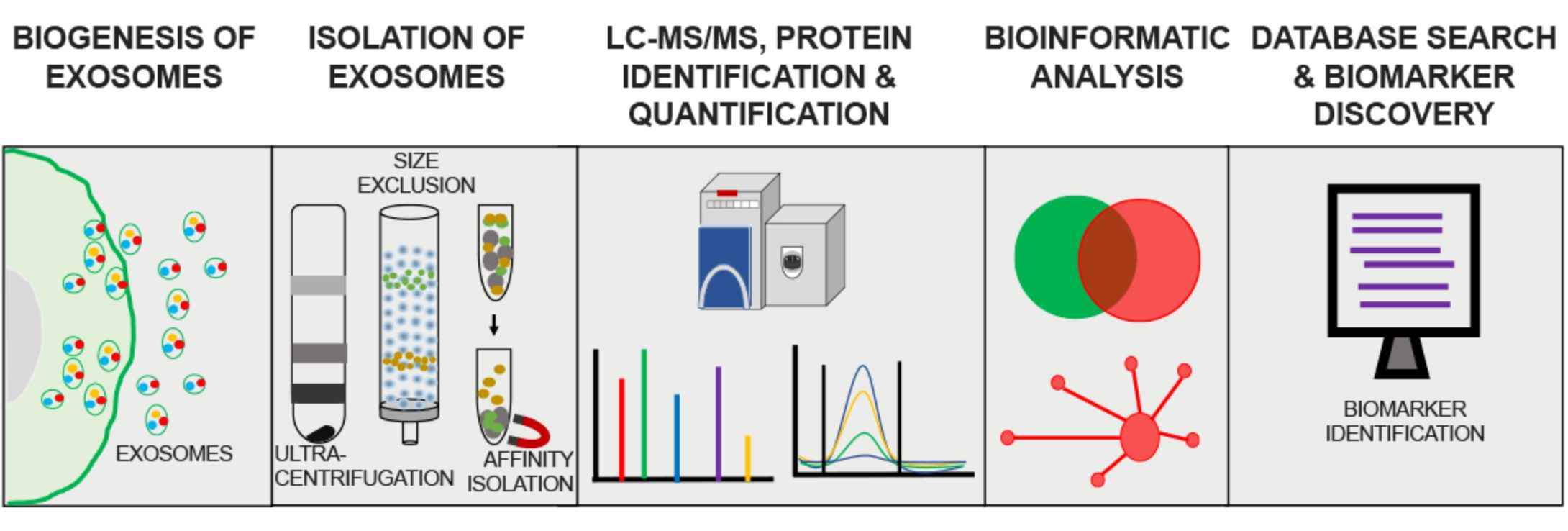 Figure 3. Process of exosome proteomics analysis. (Mathew B, et al., 2021)
Figure 3. Process of exosome proteomics analysis. (Mathew B, et al., 2021)
Lipidomics and Metabolomics Analysis of Exosomes
Exosome metabolomics, as a new force in exosome research, helps to comprehensively study the exosome internal contents and realize the analysis of the biological functions of exosomes from multiple perspectives. Lipidomics is a branch of metabolomics, aiming to analyze lipid species. Lipids play an important role in exosomes and have multiple biological functions. Dysregulation of lipid metabolism has been reported in cancer, neurodegenerative diseases, and cardiovascular diseases. Exosomes are enriched with lipids such as cholesterol, phosphatidylserine PS, phosphatidylcholine PC, and phosphatidylinositol PI, suggesting that exosomes may act as intercellular lipid mediators. From a practical and clinical point of view, the study of lipidomics and metabolomics of exosomes isolated from biofluids is the most appropriate approach as these exosomes contain disease biomarkers.
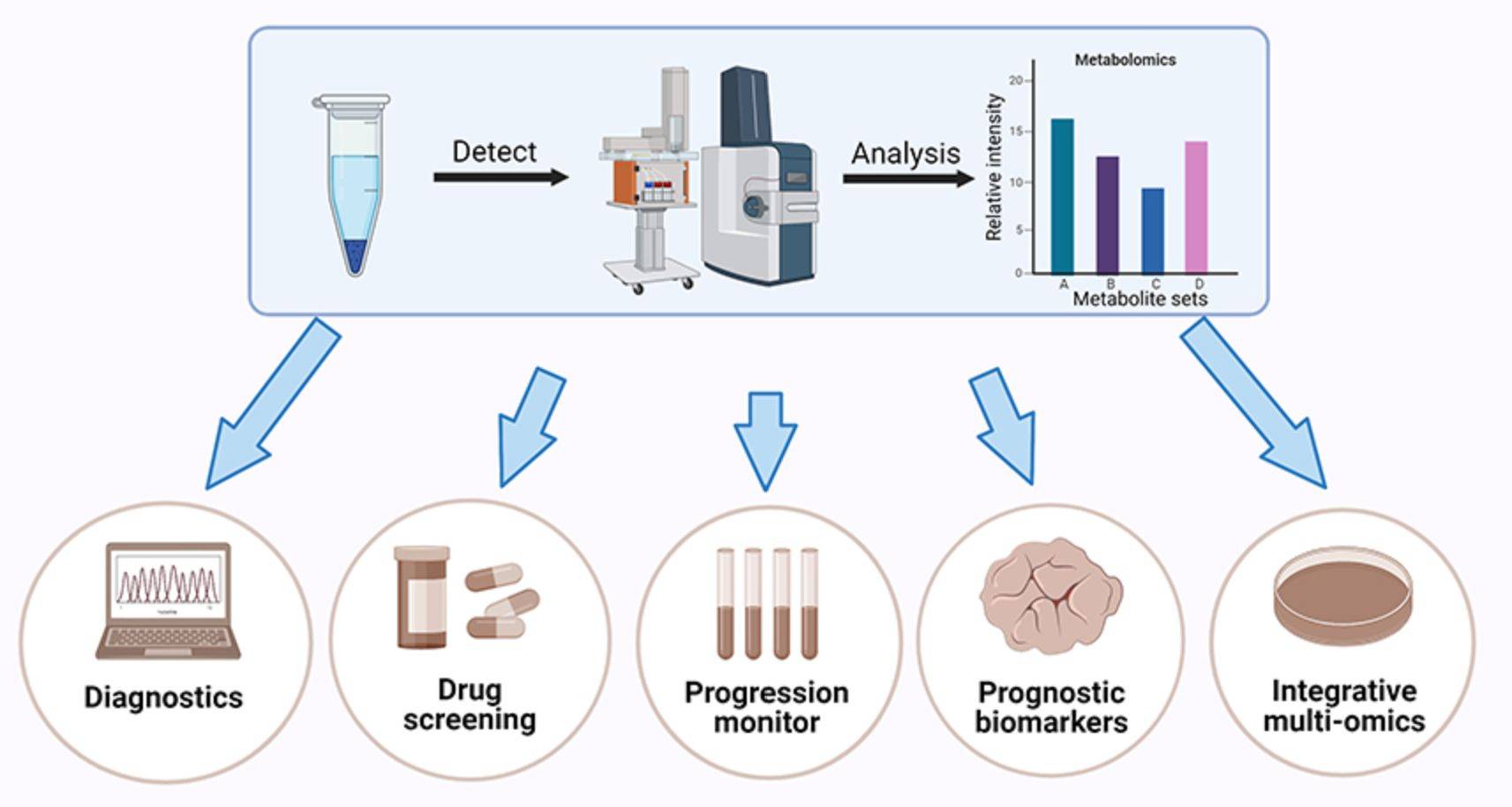 Figure 4. Applications of exosomal metabolomics. (Wu Y, et al., 2022)
Figure 4. Applications of exosomal metabolomics. (Wu Y, et al., 2022)
Transcriptomic Analysis of Exosomes
Several studies have shown that exosomes transfer proteins, lipids, and RNA between various cell types, thereby mediating intercellular communication and signaling. In particular, small non-coding RNAs in exosomes are involved in molecular events in recipient cells. In addition, RNA cargoes in exosomes have great potential to serve as non-invasive biomarkers for a wide range of diseases. In recent years, researchers have used next-generation sequencing to comprehensively analyze exosomes, characterize the amount of RNA in exosomes released by various cells, and demonstrate that miRNAs can serve as highly specific biomarkers of pathological conditions and correlate with the presence of malignant tumors.
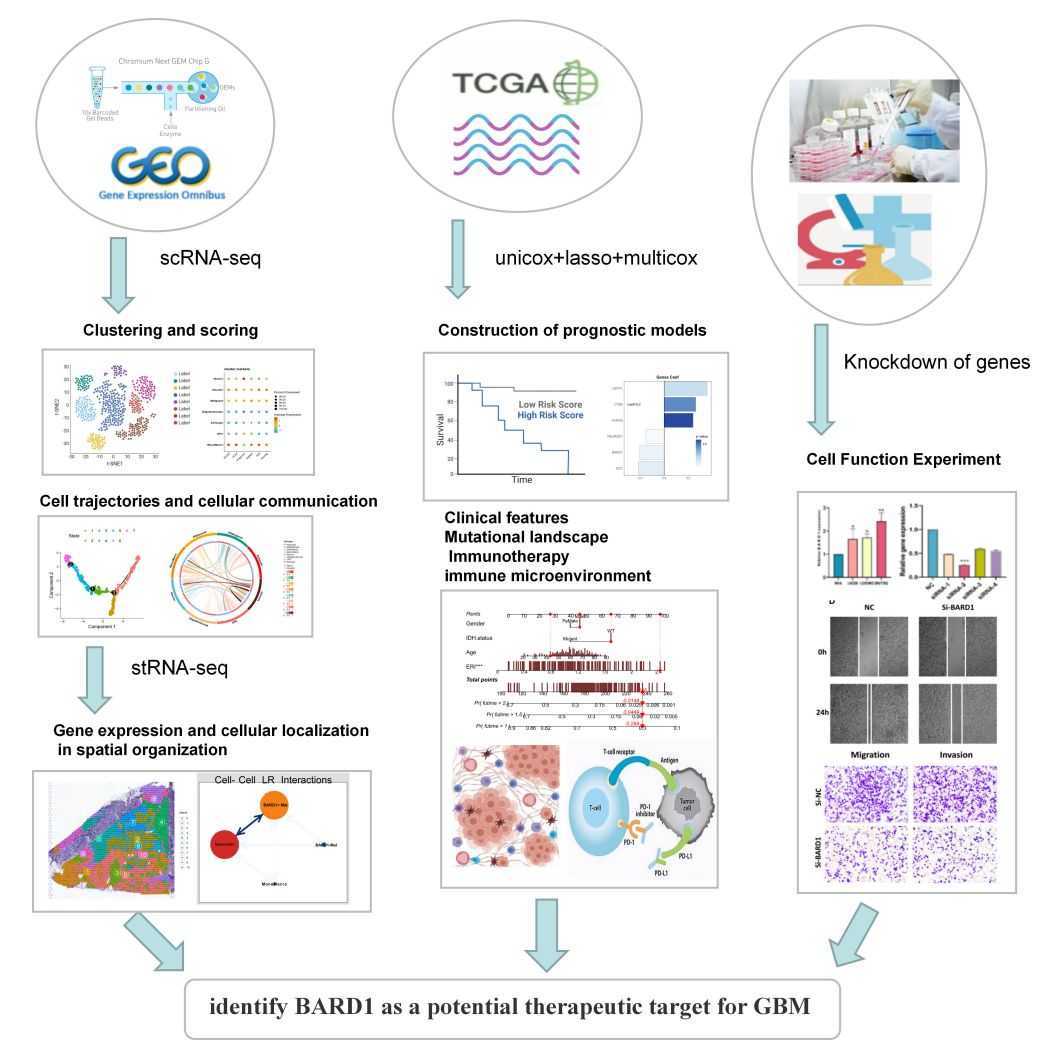 Figure 5. Using single-cell sequencing and spatial transcriptome sequencing identify glioblastoma exosome BARD1 as a potential therapeutic target. (Zhao S, et al., 2023)
Figure 5. Using single-cell sequencing and spatial transcriptome sequencing identify glioblastoma exosome BARD1 as a potential therapeutic target. (Zhao S, et al., 2023)
Our Services and Products
Creative Biostructure is a pioneering biotechnology company dedicated to advancing the field of exosome research and unlocking the therapeutic potential of these extracellular vesicles. Our renowned team of scientists and researchers specialize in comprehensive multi-omics analyses of exosomes, utilizing cutting-edge proteomics, transcriptomics, modificationomics, microbiomics, lipidomics, and metabolomics technologies to provide clients with unparalleled insights into the composition and functional capabilities of exosomes. We enable clients to accelerate drug discovery and development efforts, identify new biomarkers, and design innovative exosome-based therapeutic strategies. Please feel free to contact us for more information.
The products we offer are listed below.
| Cat No. | Product Name | Source |
| PNE-VB13 | PNExo™ Exosome-Broccoli | Exosome derived from Broccoli |
| PNE-VB14 | PNExo™ Exosome-Brussels Sprout | Exosome derived from Brussels Sprout |
| PNE-FB34 | PNExo™ Exosome-Blueberry | Exosome derived from Blueberry |
| PNE-FP93 | PNExo™ Exosome-Prickly Pear | Exosome derived from Prickly Pear |
| PNE-FLD10 | PNExo™ Exosome-Daffodil | Exosome derived from Daffodil |
| Exo-PDELN01 | HQExo™ Exosome-Garlic | Exosome derived from Garlic |
| Exo-PDELN02 | HQExo™ Exosome-Ginger | Exosome derived from Ginger |
| Exo-PDELN03 | HQExo™ Exosome-Onion | Exosome derived from Onion |
| Exo-PDELN04 | HQExo™ Exosome-Potato | Exosome derived from Potato |
| PNE-FLF63 | PNExo™ Exosome-Forget-me-not | Exosome derived from Forget-me-not |
| PNE-FLM66 | PNExo™ Exosome-Magnolia | Exosome derived from Magnolia |
| PNE-FLC68 | PNExo™ Exosome-Cockscomb | Exosome derived from Cockscomb |
| PNE-FLG70 | PNExo™ Exosome-Green Tea | Exosome derived from Green Tea |
| Exo-CH10 | HQExo™ Exosome-HCT116 | Exosome derived from human colorectal carcinoma cell line (HCT116 cell line) |
| Exo-CH11 | HQExo™ Exosome-K-562 | Exosome derived from human pleural effusion, leukemia chronic myelogenous (K-562 cell line) |
| Exo-CH16 | HQExo™ Exosome-COLO1 | Exosome derived from human colon carcinoma (COLO1 cell line) |
| Exo-CH21 | HQExo™ Exosome-A375 | Exosome derived from human malignant melanoma cell line (A375 cell line) |
| Explore All Exosome Products | ||
Creative Biostructure's team of scientists has specialized knowledge and technology to provide comprehensive exosome and related services to help clients develop exosome-based medical aesthetic products. If you have exosome research needs, please feel free to contact us for details.
References
- Lee YJ, et al. Regulation of cargo selection in exosome biogenesis and its biomedical applications in cancer. Exp Mol Med. 2024. 56(4): 877-889.
- Huang Y, et al. Recent Advances in the Use of Exosomes in Sjögren's Syndrome. Front Immunol. 2020. 11: 1509.
- Mathew B, et al. Exosomes as Emerging Biomarker Tools in Neurodegenerative and Neuropsychiatric Disorders—A Proteomics Perspective. Brain Sciences. 2021. 11(2): 258.
- Wu Y, et al. Metabolomics of Extracellular Vesicles: A Future Promise of Multiple Clinical Applications. Int J Nanomedicine. 2022. 17: 6113-6129.
- Zhao S, et al. Combining single-cell sequencing and spatial transcriptome sequencing to identify exosome-related features of glioblastoma and constructing a prognostic model to identify BARD1 as a potential therapeutic target for GBM patients. Front Immunol. 2023. 14: 1263329.