Protein Structure and Its Relationship to Function
Protein structure is central to its function, as each level of structural organization contributes to a protein's specific role in biological systems. Primary structure, the amino acid sequence, dictates the protein's folding pathways due to the unique chemical properties of each amino acid. This sequence influences the formation of local secondary structures, such as alpha helices and beta sheets, which are stabilized through hydrogen bonds. Secondary structures provide stability and framework, arranging amino acid residues in precise orientations crucial for binding or catalytic activities. When these secondary structures assemble into a compact, three-dimensional shape, they form the tertiary structure, where specific regions of the protein, such as active sites or binding domains, become functional. Some proteins operate as individual molecules, but many assemble with other proteins or subunits to form complexes, establishing a quaternary structure. This level of organization enables cooperative interactions among subunits, allowing proteins to engage in complex cellular tasks, such as signal transduction or enzymatic catalysis.
Functional Domains and Motifs in Proteins
Proteins often consist of distinct functional domains—structural units within a protein that confer specific activities. These domains can function independently or interact with other regions to carry out complex tasks. Many enzymes contain catalytic domains that facilitate biochemical reactions, along with additional binding domains that enhance substrate specificity or regulate activity. Modular domain architecture allows proteins to participate in multiple cellular pathways, adjusting their function based on cellular signals or environmental changes.
Proteins also include structural motifs, recurring patterns such as alpha helices, beta sheets, and loops, that contribute to their functional roles. Coiled-coils, for example, are a motif where alpha helices wrap around each other, providing a stable interface for protein-protein interactions. Similarly, beta-barrel structures are often found in membrane proteins, where they form pores or channels for transporting ions and small molecules. These motifs highlight the connection between structure and function, as small structural features can facilitate specific biological processes, like molecular transport or signal transmission.
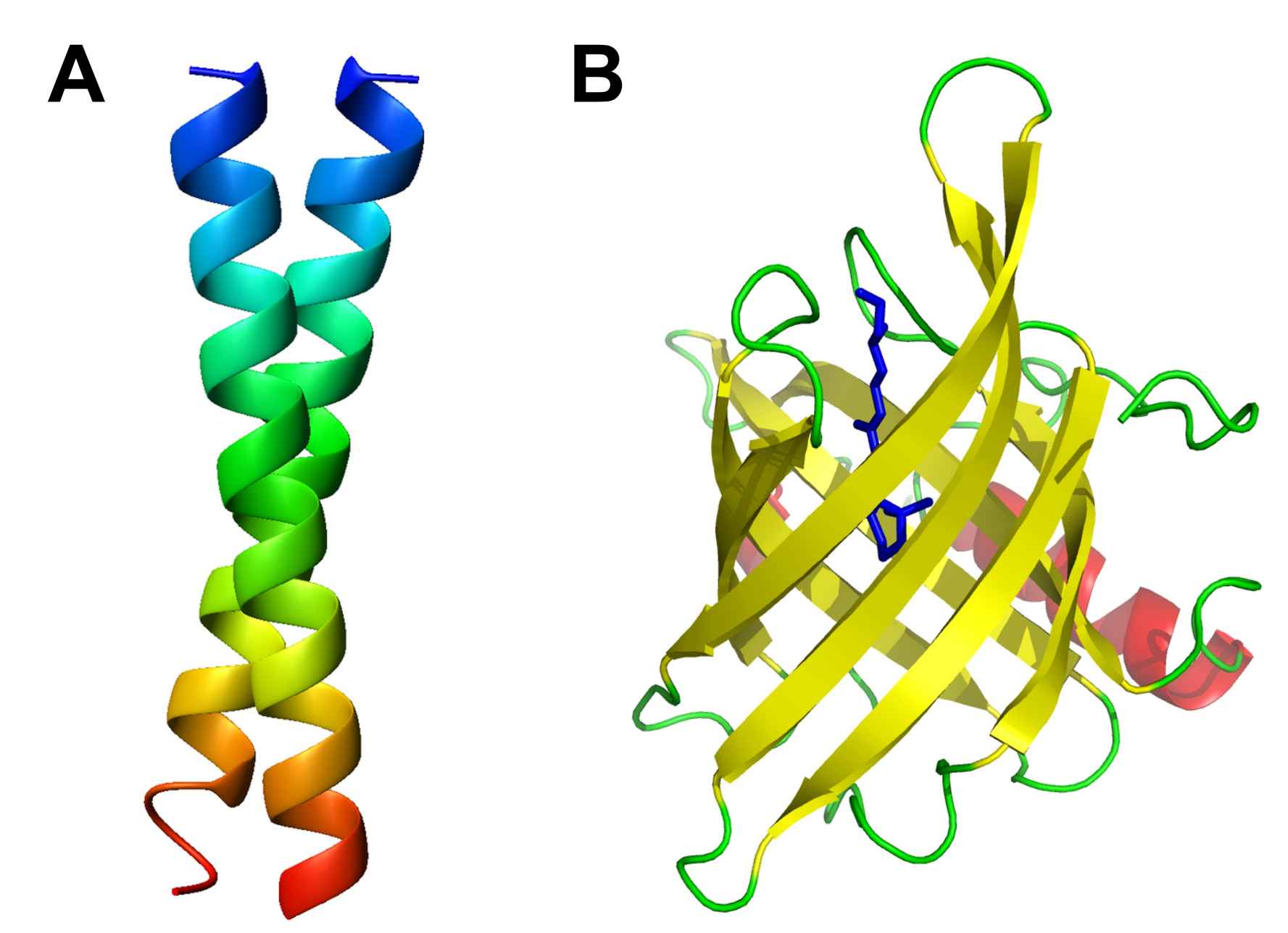 Figure 1: A. The classic example of a coiled coil is the GCN4 leucine zipper (PDB: 1ZIK). B. 8-strand β barrel. Human retinol-binding protein bound to retinol (vitamin A) in blue (PDB: 1RBP).
Figure 1: A. The classic example of a coiled coil is the GCN4 leucine zipper (PDB: 1ZIK). B. 8-strand β barrel. Human retinol-binding protein bound to retinol (vitamin A) in blue (PDB: 1RBP).
Enzyme Function and Catalytic Mechanisms
Enzymes, a class of proteins essential for catalyzing biochemical reactions, are an example of how protein structure facilitates function. Each enzyme contains an active site—a specialized pocket formed by the tertiary structure that binds substrates with high specificity. The shape and chemical environment of the active site lower the activation energy required for a reaction, accelerating reaction rates and enabling processes that would otherwise be too slow for cellular life.
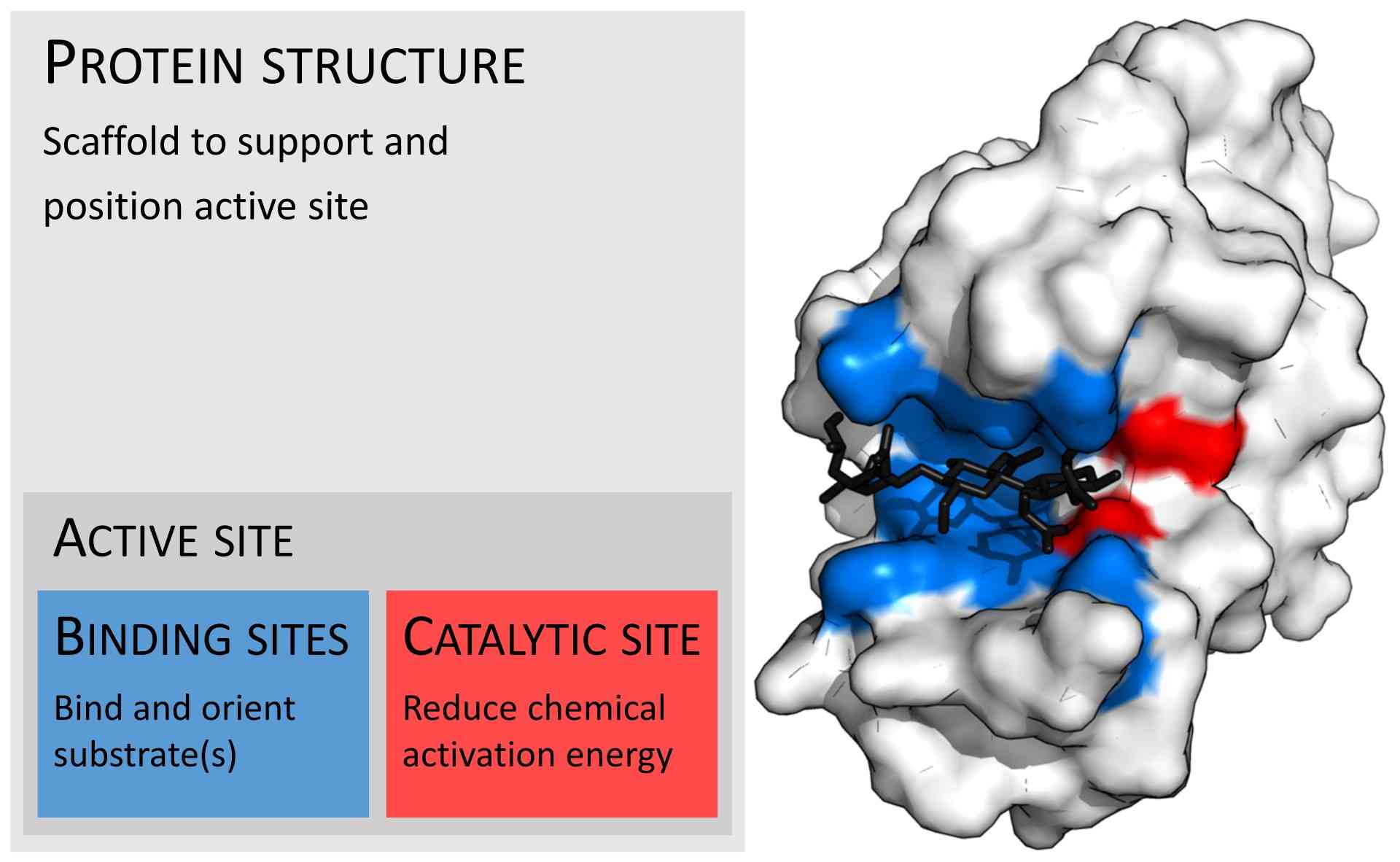 Figure 2: Organization of enzyme structure and lysozyme example. Binding sites in blue, catalytic site in red and peptidoglycan substrate in black (PDB: 9LYZ).
Figure 2: Organization of enzyme structure and lysozyme example. Binding sites in blue, catalytic site in red and peptidoglycan substrate in black (PDB: 9LYZ).
Structural biology plays a key role in uncovering how enzymes work at a molecular level. Take lysozyme, for example. This enzyme's active site is arranged with amino acid residues that interact with its target in specific orientations to break down bacterial cell walls. Another example is DNA polymerase, which ensures the accurate pairing of nucleotides during DNA replication owe to its precisely organized active site.
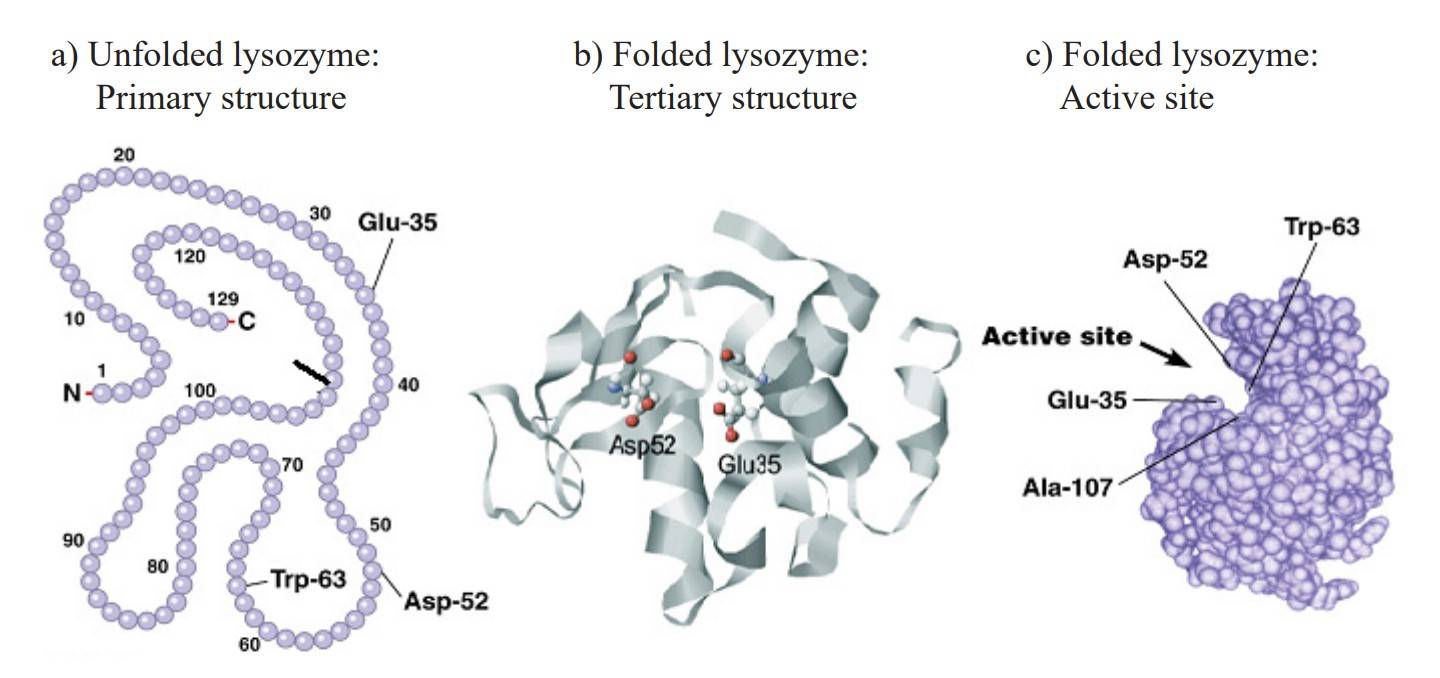 Figure 3: Chicken lysozyme primary (a) and tertiary structure (b). Active site with the amino acids responsible for the lysozyme activity (c). (Gálvez-Iriqui et al., 2020)
Figure 3: Chicken lysozyme primary (a) and tertiary structure (b). Active site with the amino acids responsible for the lysozyme activity (c). (Gálvez-Iriqui et al., 2020)
Protein Interactions and Complexes
Proteins don't usually work alone. Protein interactions are ubiquitous in living organisms. Thanks to advances in structural biology, scientists have been able to uncover how these protein complexes are organized and how their interactions help the cell function. An example is the ribosome. This massive structure, made of proteins and RNA, is responsible for building new proteins in the cell. Using cryo-EM, researchers have mapped out the ribosome in incredible detail. These studies show how its proteins and RNA components work together to translate genetic information into functional proteins. It's a fascinating glimpse into how teamwork at the molecular level powers life!
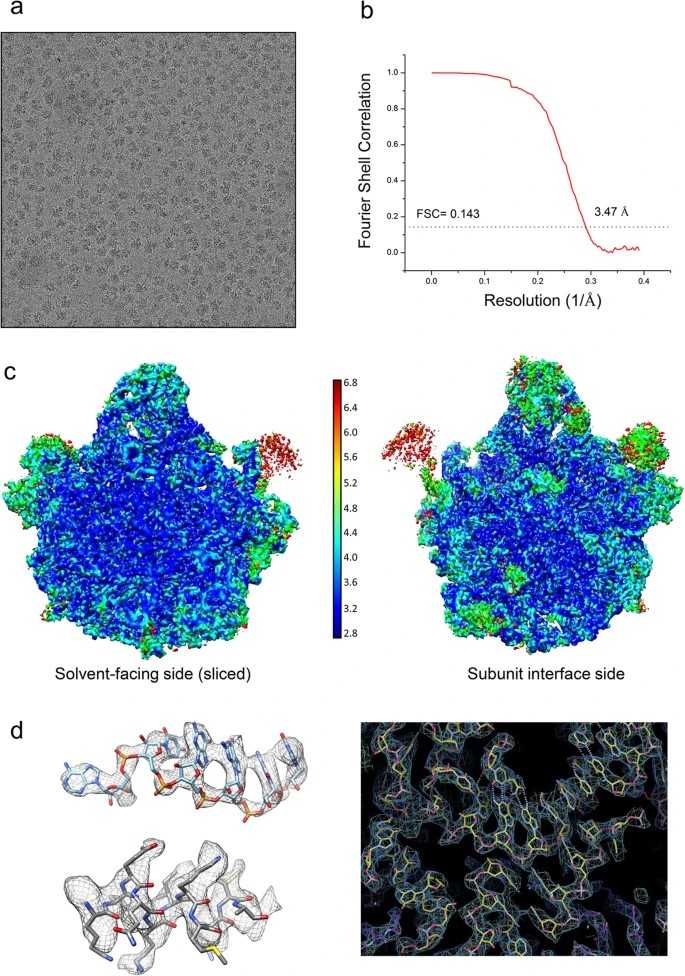 Figure 4: Studying the large subunit of the spinach chloroplast ribosome using Cryo-EM. (a) Representative micrograph showing ribosome particles. (b) Fourier Shell Correlation (FSC) curve showing the average resolution of the reconstructed density map of the spinach chloro-ribosome LSU. (c) Local resolution estimation of cryo-EM map by ResMap. Map is colored according to local resolution of masked map. (d) Representative density showing a 23S rRNA region (nucleotides 2584–2589) in upper left, a helix of PSRP5 protein (amino acid residues 99–109) in lower left and a region around nucleotide G2462 in 23S rRNA on the right, showing quality of the map and fitting of the models. (Ahmed et al., 2016)
Figure 4: Studying the large subunit of the spinach chloroplast ribosome using Cryo-EM. (a) Representative micrograph showing ribosome particles. (b) Fourier Shell Correlation (FSC) curve showing the average resolution of the reconstructed density map of the spinach chloro-ribosome LSU. (c) Local resolution estimation of cryo-EM map by ResMap. Map is colored according to local resolution of masked map. (d) Representative density showing a 23S rRNA region (nucleotides 2584–2589) in upper left, a helix of PSRP5 protein (amino acid residues 99–109) in lower left and a region around nucleotide G2462 in 23S rRNA on the right, showing quality of the map and fitting of the models. (Ahmed et al., 2016)
The proteasome is another fascinating example of how proteins work together in complexes. This complex operates as a molecular machine, selectively binding to ubiquitin-tagged proteins and breaking them down into smaller peptides. By studying the structure of the proteasome, scientists have uncovered how it manages protein recycling. This process, known as protein turnover, is vital for maintaining a healthy, balanced cell environment.
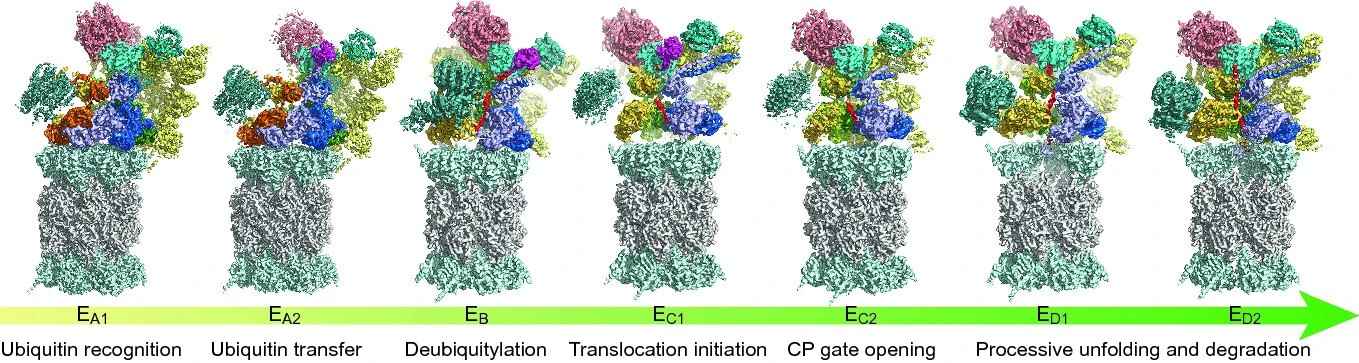 Figure 5: Seven cryo-EM maps of the substrate-engaged Homo sapiens 26S proteasome at 2.8–3.6 Å resolution capture the key intermediate steps of substrate processing and provide important insights into the chemical cycle of proteasome-mediated degradation. (Dong et al., 2019)
Figure 5: Seven cryo-EM maps of the substrate-engaged Homo sapiens 26S proteasome at 2.8–3.6 Å resolution capture the key intermediate steps of substrate processing and provide important insights into the chemical cycle of proteasome-mediated degradation. (Dong et al., 2019)
Membrane Proteins and Signal Transduction
Membrane proteins are essential for many important cell functions, like signal transduction, transporting molecules, and cellular communication. Structural biology has provided insights into the architecture of these proteins, explaining how they function within the lipid bilayer. For example, a large protein family—G-protein-coupled receptors (GPCRs) detect signals from outside the cell and trigger responses inside the cell. Structural studies of GPCRs have shown how these proteins change conformation upon binding ligands, triggering downstream signaling cascades that regulate diverse physiological processes.
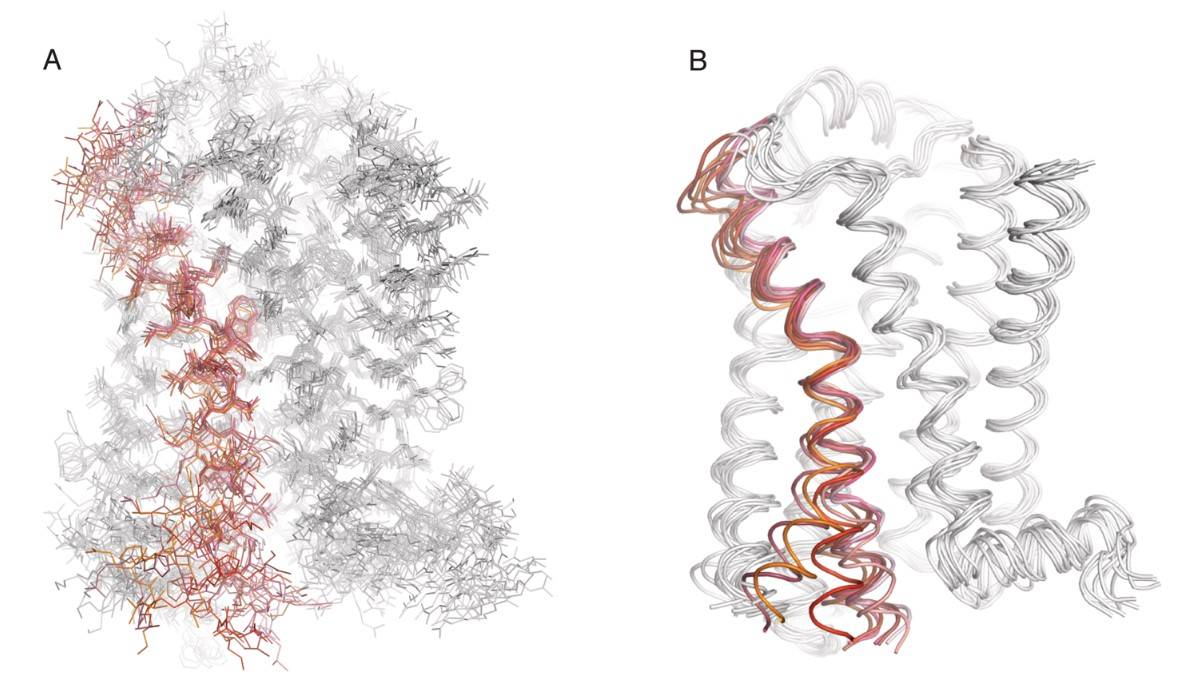 Figure 6. Atomic-level motions of a GPCR revealed through MD simulations. Representative frames from MD simulations (from Dror et al., 2011) of agonist-bound β2AR as it transitions from an active state to an inactive state, with (A) all non-hydrogen atoms represented as lines and (B) protein backbone represented as ribbons. Transmembrane helix 6 (TM6) is colored red to highlight its high degree of mobility during the transition between active and inactive states. (Latorraca et al., 2017)
Figure 6. Atomic-level motions of a GPCR revealed through MD simulations. Representative frames from MD simulations (from Dror et al., 2011) of agonist-bound β2AR as it transitions from an active state to an inactive state, with (A) all non-hydrogen atoms represented as lines and (B) protein backbone represented as ribbons. Transmembrane helix 6 (TM6) is colored red to highlight its high degree of mobility during the transition between active and inactive states. (Latorraca et al., 2017)
Ion channel proteins are another important example. They control the flow of ions across cell membranes, which is key for electrical signaling in nerve and muscle cells. Structural studies have revealed the gating mechanisms of ion channels, showing how they open or close in response to voltage changes or ligand binding. These insights are crucial for understanding how our nervous and muscular systems work. They also help in developing new drugs that can target these pathways to treat various conditions.
Methods of Studying Protein Structure
Understanding how proteins function starts with uncovering their three-dimensional structures. Several experimental techniques in structural biology make this possible.
X-ray crystallography is a cornerstone of structural research. By analyzing how X-rays interact with protein crystals, this method provides detailed, atomic-level images. It has been instrumental in revealing the structures of proteins ranging from small enzymes to complex molecular machines.
Nuclear Magnetic Resonance (NMR) spectroscopy offers another valuable approach. This technique is particularly valuable for studying proteins in solution, giving researchers the ability to observe them in environments that closely mimic their natural state. NMR is ideal for exploring dynamic processes like protein folding, ligand interactions, and structural flexibility.
Cryo-Electron Microscopy (cryo-EM) has revolutionized structural biology with its ability to visualize large and complex structures, such as viruses or protein assemblies, without requiring crystallization. By freezing proteins in thin layers of ice, cryo-EM preserves their near-native shapes, providing a closer look at molecular details that drive cellular processes.
Together with computational modeling tools, these techniques enable scientists to connect protein structures to their functions, deepening our understanding of biological systems.
In short, protein function is intrinsically linked to its structure, and structural biology provides the tools to unlock this important connection. By exploring the detailed architecture of proteins, structural biology has transformed our understanding of protein interactions, enzymatic catalysis, and cellular signaling pathways. These insights not only enhance our knowledge of basic biology but also have practical applications in areas like drug discovery and biotechnology.
At Creative Biostructure, we specialize in protein structure analysis, offering a range of high-quality services and customized solutions, including X-ray crystallography, Cryo-EM, and other analysis methods. With our extensive expertise and cutting-edge equipment, we use advanced techniques to provide accurate, detailed insights into protein function. Choose Creative Biostructure to drive your protein function studies forward, contact us now!
References
- Ahmed T, Yin Z, Bhushan S. Cryo-EM structure of the large subunit of the spinach chloroplast ribosome. Sci Rep. 2016;6(1):35793.
- Dong Y, Zhang S, Wu Z, et al. Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome. Nature. 2019;565(7737):49-55.
- Dror RO, Arlow DH, Maragakis P, et al. Activation mechanism of the β2 -adrenergic receptor. Proc Natl Acad Sci USA. 2011;108(46):18684-18689.
- Gálvez-Iriqui AC, Plascencia-Jatomea M, Bautista-Baños S. Lysozymes: characteristics, mechanism of action and technological applications on the control of pathogenic microorganisms. RMF. 2020;38(3).
- Latorraca NR, Venkatakrishnan AJ, Dror RO. GPCR dynamics: structures in motion. Chem Rev. 2017;117(1):139-155.
- Mao Y. Structure, dynamics and function of the 26s proteasome. In: Harris JR, Marles-Wright J, eds. Macromolecular Protein Complexes III: Structure and Function. Springer International Publishing; 2021:1-151.