Structural Research of Vascular Endothelial Growth Factor Receptors
Vascular endothelial growth factor (VEGF) binds to a variety of membrane receptors (VEGFR) to regulate vertebrate biological responses, including vasculogenesis, vascular development, and lymphangiogenesis. VEGFR is a widely researched membrane-bound receptor tyrosine kinase protein, of which VEGFR-1 is required for hematopoietic stem cell recruitment as well as monocyte and macrophage migration, VEGFR-2 regulates vascular endothelial function, and VEGFR-3 regulates lymphatic endothelial function. In recent years, research has revealed that VEGF-VEGFR dysregulation is involved in many diseases, especially cancer. Therefore, research on VEGF-VEGFR will help to develop therapeutic approaches for such diseases.
Overall structural analysis of VEGFR
The kinase structural domains of VEGFR are conserved and have high sequence identity. Most VEGFRs display similar primary structures, including seven immunoglobulin (Ig)-like domains, a short α-helical transmembrane domain, cytoplasmic near-membrane regions, and a tyrosine kinase (TK) domain linked to a flexible carboxyl-terminal linkage. The binding of VEGFRs to homologous ligands triggers the formation of homodimers, which activate cellular signaling pathways that lead to a wide range of cellular functions.
Advances in structural research of VEGF-VEGFR complexes
To date, many structural studies of the VEGF/VEGFR complex based on single-particle electron microscopy, small-angle X-ray scattering, and X-ray crystallography have shown how the ligand binds to the Ig structural domain at the distal end of the membrane. The structures show the presence of two 1:1 complexes in the asymmetric unit and two receptors linked by the dimeric VEGF-A bound to the Ig structural domain. The research also shows that homotypic receptor-receptor contacts in Ig structural domains 4-7 increase the binding affinity of VEGFR-1 ECD to VEGF-A.
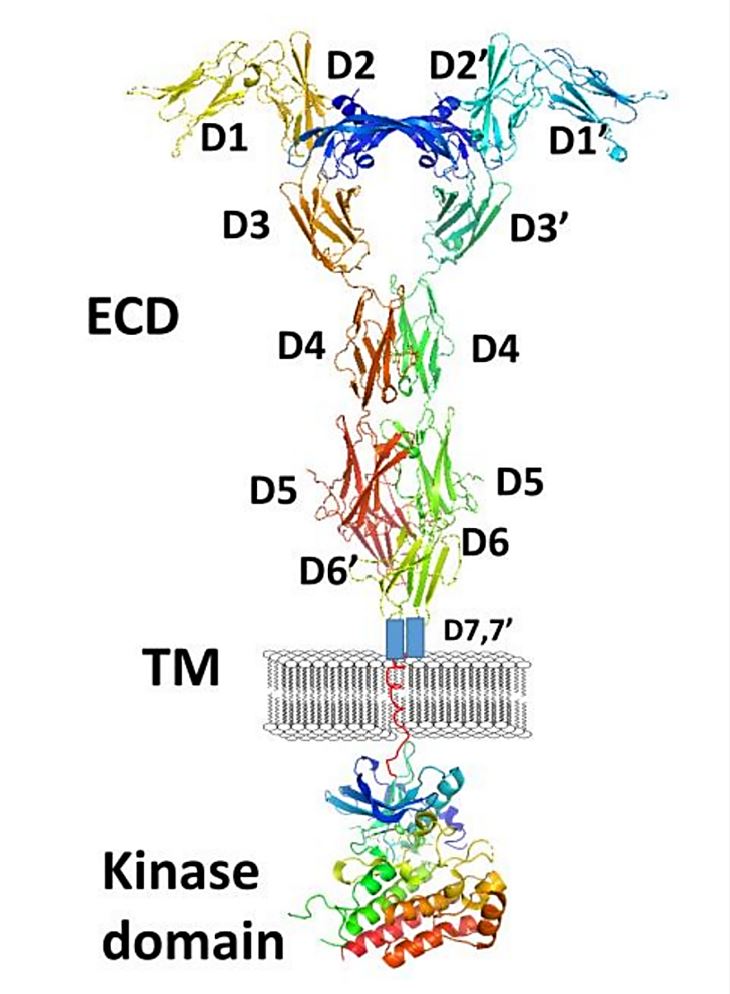 Figure 1. A schematic representation of full-length VEGFR-1 with VEGF-A. (Kaufman NEM, et al., 2021)
Figure 1. A schematic representation of full-length VEGFR-1 with VEGF-A. (Kaufman NEM, et al., 2021)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Dimeric G-quadruplex DNA formed in the proximal promoter of VEGFR-2 | DNA molecule | SOLUTION NMR | / | 7XH3 |
| Dimeric G-quadruplex DNA formed in the proximal promoter of VEGFR-2 | synthetic construct | SOLUTION NMR | / | 7XFV |
| G-quadruplex formed in vegfr-2 proximal promoter sequence | synthetic construct | SOLUTION NMR | / | 5ZEV |
| VEGF-A in complex with VEGFR-1 domains D1-6 | Homo sapiens | X-ray diffraction | 4 Å | 5T89 |
| VEGFR-1 domain 2 in presence of Zn | Homo sapiens | X-ray diffraction | 2.055 Å | 4CKV |
| VEGFR-3 extracellular domains D4-5 | Homo sapiens | X-ray diffraction | 2.5 Å | 4BSJ |
| VEGFR-1 domain 2 in presence of Cobalt | Homo sapiens | X-ray diffraction | 2 Å | 4CL7 |
| VEGFR-1 domain 2 in presence of CU | Homo sapiens | X-ray diffraction | 1.995 Å | 5ABD |
| VEGFR-2/VEGF-E complex | Homo sapiens | X-ray diffraction | 3.205 Å | 3V6B |
| VEGF-C in complex with VEGFR-3 domains D1-2 | Homo sapiens | X-ray diffraction | 4.201 Å | 4BSK |
| The Binding of VEGF-B by VEGFR-1D2 | Homo sapiens | X-ray diffraction | 2.71 Å | 2XAC |
| The trimeric mutant TM domain of VEGFR2 receptor | Homo sapiens | SOLUTION NMR | / | 2MET |
| The VEGFR2 kinase domain in complex with axitinib (AG-013736) | Homo sapiens | X-ray diffraction | 1.95 Å | 4AG8 |
| VEGFR2 kinase domain in complex with PF- 00337210 | Homo sapiens | X-ray diffraction | 1.5 Å | 2XIR |
| VEGFR2 (Juxtamembrane and Kinase Domains) in complex with sorafenib (BAY 43-9006) | Homo sapiens | X-ray diffraction | 2.03 Å | 4ASO |
| VEGFR2 in complex with a 3,4,5-trimethoxy aniline containing pyrimidine | Homo sapiens | X-ray diffraction | 2.15 Å | 3CJF |
| VEGFR IN complex with tivozanib (AV-951) | Homo sapiens | X-ray diffraction | 1.83 Å | 4ASE |
| VEGFR1 in complex with N-(4-Chlorophenyl)-2-((pyridin-4-ylmethyl) amino) benzamide | Homo sapiens | X-ray diffraction | 2.7 Å | 3HNG |
| The dimeric VEGFR2 membrane domain | Homo sapiens | SOLUTION NMR | / | 2M59 |
| The mutant dimeric TM domain of VEGFR2 receptor | Homo sapiens | SOLUTION NMR | / | 2MEU |
| Vegfr2 with a benzimidazole-urea inhibitor | Homo sapiens | X-ray diffraction | 2.05 Å | 2OH4 |
| PlGF in complex with domain 2 of VEGFR1 | Homo sapiens | X-ray diffraction | 2.45 Å | 1RV6 |
| VEGFR-2 domains 4-5 in complex with DARPin D4b | Homo sapiens | X-ray diffraction | 2.38 Å | 5OYJ |
Table 1. Structural research of the vascular endothelial growth factor receptors.
Understanding the structure and function of membrane proteins, such as vascular endothelial growth factor receptors, is essential for the development of new drugs and therapies for the treatment of related diseases, and Creative Biostructure is a leading provider of structural analysis services for membrane proteins. Our team of experts uses X-ray crystallography, cryo-electron microscopy (cryo-EM), and NMR spectroscopy to determine the three-dimensional structure of membrane proteins and understand their function.
If you are interested in gaining a deeper understanding of the structure of membrane proteins and other biomolecules, our team of experts is the right choice. With many years of experience and a wealth of knowledge, we are always available to discuss your research requirements and provide you with the best solution for your project. Contact us today to learn more about our comprehensive structural analysis services and how we can help take your research to the next level.
References
- Kaufman NEM, et al. Molecular Targeting of Epidermal Growth Factor Receptor (EGFR) and Vascular Endothelial Growth Factor Receptor (VEGFR). Molecules. 2021. 26(4): 1076.
- Holmes K, et al. Vascular endothelial growth factor receptor-2: structure, function, intracellular signaling, and therapeutic inhibition. Cell Signal. 2007. 19(10): 2003-2012.
- Park SA, et al. Structure and function of vascular endothelial growth factor and its receptor system. BMB Rep. 2018. 51(2): 73-78.
- Shaik F, et al. Structural Basis for Vascular Endothelial Growth Factor Receptor Activation and Implications for Disease Therapy. Biomolecules. 2020. 10(12): 1673.