Structural Research of Aquaporins and Glyceroporins
Aquaporins and glyceroporins are a class of transmembrane proteins that serve as gatekeepers for the transport of water and glycerol across cell membranes. These proteins have a critical role in regulating the water and energy balance within the body, and their dysfunction has been linked to an array of diseases, ranging from diabetes to kidney disorders.
The structure of aquaporin-1 (AQP1) was first determined in 1994, illuminating a water-selective channel that allows the rapid transport of water molecules across the membrane. A recent study used X-ray crystallography to determine the structure of human aquaglyceroporin AQP7 in complex with glycerol. The results revealed that AQP7 has a distinct architecture that grants it the ability to selectively transport glycerol, while simultaneously excluding other solutes. The study also found that AQP7 undergoes conformational changes during glycerol transport, which further reinforces its high selectivity. The structure of AQP7 has since been used as a template for the determination of the structures of other glyceroporins, such as AQP3 and AQP9.
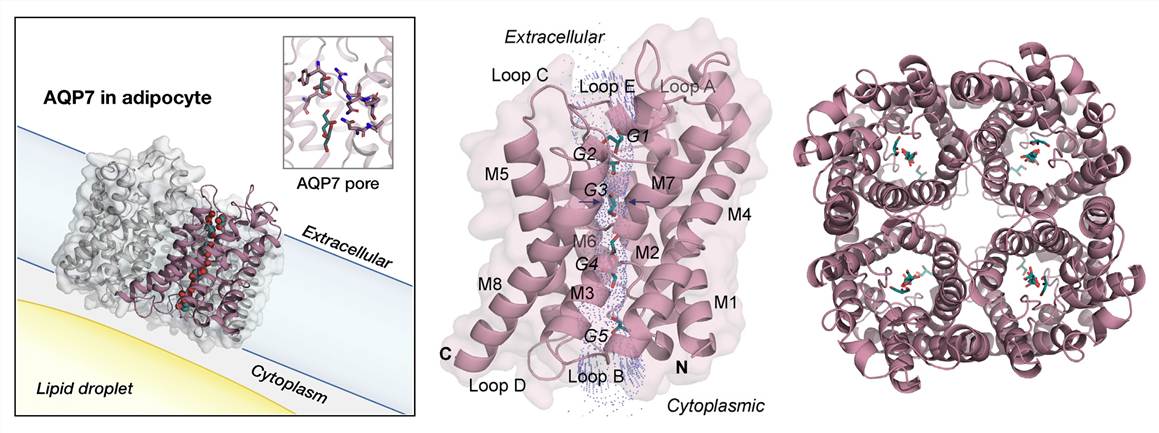 Figure 1. The overall structure of human AQP7. (de Mare S W, et al., 2020)
Figure 1. The overall structure of human AQP7. (de Mare S W, et al., 2020)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| AQP0 aquaporin water channel | Bos taurus | X-ray diffraction | 2.24 Å | 1YMG |
| AQP0 aquaporin water channel from sheep lens | Ovis aries | Electron crystallography | 3.00 Å | 1SOR |
| AQP0 aquaporin sheep lens junction | Ovis aries | Electron crystallography | 1.90 Å | 2B6O |
| AQP0 aquaporin sheep lens junction in E. coli polar lipids | Ovis aries | Electron crystallography | 2.50 Å | 3M9I |
| AQP1 red blood cell aquaporin water channel | Homo sapiens | Electron crystallography | 3.80 Å | 1FQY |
| AQP1 red blood cell aquaporin water channel | Homo sapiens | Electron crystallography | 3.70 Å | 1IH5 |
| AQP1 red blood cell aquaporin water channel (expressed in Spodoptera frugiperda) | Homo sapiens | X-ray diffraction | 3.28 Å | 4CSK |
| AQP1 red blood cell aquaporin water channel (expressed in Komagataella pastoris) | Homo sapiens | Solid-state NMR | / | 6POJ |
| AQP1 aquaporin red blood cell water channel | Bos taurus | X-ray diffraction | 2.20 Å | 1J4N |
| AQP2 Aquaporin from kidney (expressed in Komagataella phaffii) | Homo sapiens | X-ray diffraction | 2.75 Å | 4NEF |
| AQP2 Aquaporin from kidney crystallized on a silicon chip (expressed in Komagataella phaffii) | Homo sapiens | X-ray diffraction | 3.70 Å | 6QF5 |
| AQP4 aquaporin rat glial cell water channel (expressed in Spodoptera frugiperda) | Rattus norvegicus | Electron crystallography | 3.70 Å | 2D57 |
| AQP4 aquaporin rat glial cell water channel (expressed in Spodoptera frugiperda) | Rattus norvegicus | Electron crystallography | 2.80 Å | 2ZZ9 |
| AQP4 aquaporin water channel (expressed in Komagataella pastoris) | Homo sapiens | X-ray diffraction | 1.80 Å | 3GD8 |
| Aquaporin 5 (AQP5) (expressed in Komagataella pastoris) | Homo sapiens | X-ray diffraction | 2.00 Å | 3D9S |
| Aquaporin 7 (expressed in Komagataella pastoris) | Homo sapiens | X-ray diffraction | 1.90 Å | 6QZI |
| AQP7 dimer of tetramers_D4 (expressed in Komagataella pastoris) | Homo sapiens | Cryo-EM single particle analysis | 2.55 Å | 8AMX |
| AQP10 (expressed in Saccharomyces cerevisiae) | Homo sapiens | X-ray diffraction | 2.30 Å | 6F7H |
| AqpM aquaporin water channel (expressed in E. coli) | Methanothermobacter marburgensis | X-ray diffraction | 1.68 Å | 2F2B |
| Aquaporin Z (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.50 Å | 1RC2 |
| AqpZ aquaporin showing two conformations of Arg-189 (expressed in E. coli) | Escherichia coli | X-ray diffraction | 3.20 Å | 2ABM |
| AqpZ aquaporin (C9S/C20S), T183C mutant without Hg (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.30 Å | 2O9D |
| AqpZ mutant F43W (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.40 Å | 3NK5 |
| PIP2;1 plant aquaporin (closed conformation) (expressed in Komagataella pastoris) | Spinacia oleracea | X-ray diffraction | 2.10 Å | 1Z98 |
| PIP2;1 plant aquaporin, S115E mutant (expressed in Komagataella pastoris) | Spinacia oleracea | X-ray diffraction | 2.30 Å | 3CLL |
| PIP2;4 plant aquaporin (expressed in Komagataella pastoris) | Arabidopsis thaliana | X-ray diffraction | 3.70 Å | 6QIM |
| TIP2;1 ammonia-permeable aquaporin (expressed in Komagataella pastoris) | Arabidopsis thaliana | X-ray diffraction | 1.18 Å | 5I32 |
| GlpF glycerol facilitator channel (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.20 Å | 1FX8 |
| GlpF glycerol facilitator channel, W84F/F200T-mutant (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.10 Å | 1LDF |
| AQP aquaglyceroporin (expressed in E. coli) | Plasmodium falciparum | X-ray diffraction | 2.05 Å | 3C02 |
| Aqy1 yeast aquaporin (pH 3.5) | Komagataella pastoris | X-ray diffraction | 1.15 Å | 2W2E |
| Aqy1 yeast aquaporin (pH 8.0) (expressed in Komagataella pastoris) | Komagataella pastoris | X-ray diffraction | 0.88 Å | 3ZOJ |
| NIP2;1 aquaporin (metalloid porin) silicon transporter (expressed in Saccharomyces cerevisiae) | Oryza sativa | X-ray diffraction | 3.00 Å | 7NL4 |
| Lsi1 aquaporin (metalloid porin) silicon transporter (expressed in Spodoptera frugiperda) | Oryza sativa | X-ray diffraction | 1.80 Å | 7CJS |
Table 1. Structural Research of Aquaporins and Glyceroporins.
At Creative Biostructure, we have a team of experts with extensive experience in the structural analysis of proteins, including aquaporins and glyceroporins. We use cutting-edge techniques to determine the structure of these proteins at high resolution, providing insights into their function and potential therapeutic applications.
Our services include X-ray crystallography, cryo-electron microscopy, and nuclear magnetic resonance spectroscopy, allowing us to provide our clients with a comprehensive structural analysis of their proteins. We work closely with our clients to ensure that our services meet their specific research needs, and we provide detailed reports and analyses to help them interpret the results. If you are interested in learning more about our services, please do not hesitate to contact us directly. Our team of experts is always available to discuss your research needs and provide you with the best possible solutions.
References
- de Mare S W, et al. Structural basis for glycerol efflux and selectivity of human aquaporin 7. Structure. 2020, 28(2): 215-222. e3.
- Saitoh Y, et al. Structural basis for high selectivity of a rice silicon channel Lsi1. Nature Communications. 2021, 12(1): 6236.
- Huang P, et al. Cryo-EM structure supports a role of AQP7 as a junction protein. Nature Communications. 2023, 14(1): 600.
