RNA Structure
Ribonucleic acid (abbreviated as RNA) is similar to deoxyribonucleic acid (abbreviated as DNA) and is another type of nucleic acid. From a chemical point of view, the difference between DNA and RNA is not obvious. DNA is constructed from deoxyribonucleotides, while RNA is made from ribonucleotides (containing ribose, nitrogenous bases, and phosphate groups). The difference between the pentose sugars of them is that an oxygen atom is added to the 3' carbon atom of the ribose. Another difference is the choice of bases: RNA utilizes uracil (U) instead of thymine (T), which contains a methyl group. Additionally, RNA does not have a complementary strand and often exists in a single-stranded form. Although RNA is single-stranded, most types of RNA molecules display extensive intramolecular base pairing between complementary sequences within the RNA strand, creating a predictable three-dimensional structure for their function. Although RNA is very similar in chemical structure to DNA, its three-dimensional structure is very different.
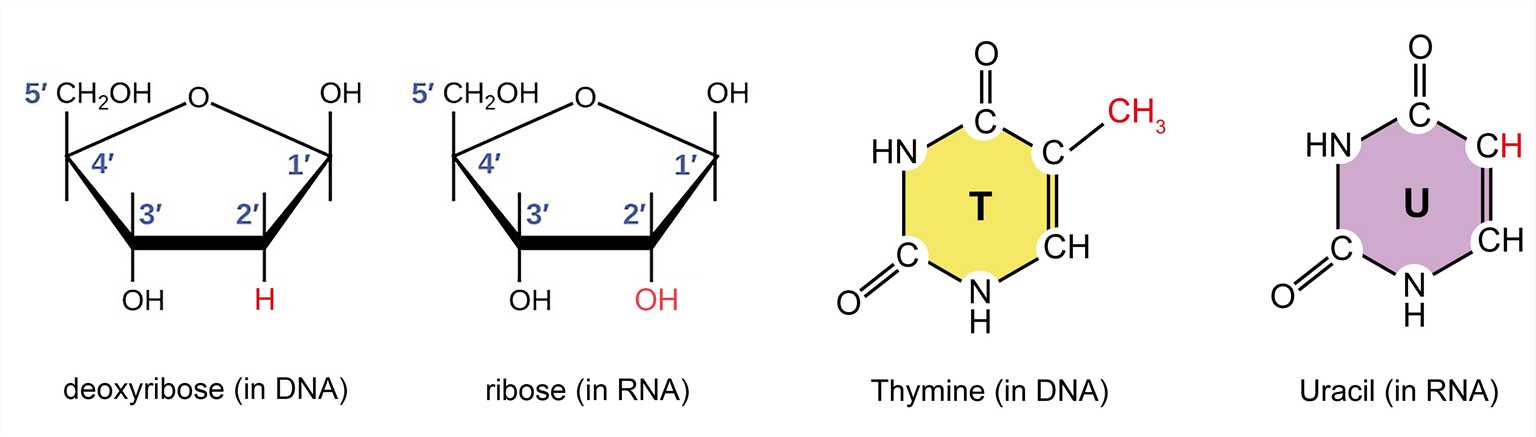 Figure 1. The difference between the pentose and nitrogenous bases of DNA and RNA.
Figure 1. The difference between the pentose and nitrogenous bases of DNA and RNA.
Levels of RNA structure
We can talk about different levels of RNA structure as we discuss protein structure. The primary structure of a nucleic acid refers to its base sequence. In RNA, the secondary structures are the two-dimensional base-pair foldings in which local sequences have regions of self-complementarity, resulting in base pairs and turns. Therefore, the secondary structure of RNA is determined by base pairing. The most complex structural level is the tertiary structure. The tertiary structure is mainly the spatial arrangement of secondary structural units. One of the things that makes sense in the molecular structure of RNA is the presence of base pairs that interact through the non-Watson-Crick base pairing principle. These base pairs are often involved in the formation and stabilization of tertiary structures. A coil can form a hydrogen bond with another part of the structure to stabilize it. In addition, van der Waals forces and electrostatic forces also contribute greatly to the tertiary structure, and finally the protein molecules can interact with and stabilize the tertiary structure of RNA. Only a limited amount of information has previously been available on the structure of RNA because of the experimental difficulties encountered in RNA crystallization. However, with the success of the ribosome crystal structure analysis, this milestone has brought a series of achievements in the high-resolution crystal structure of RNA molecules.
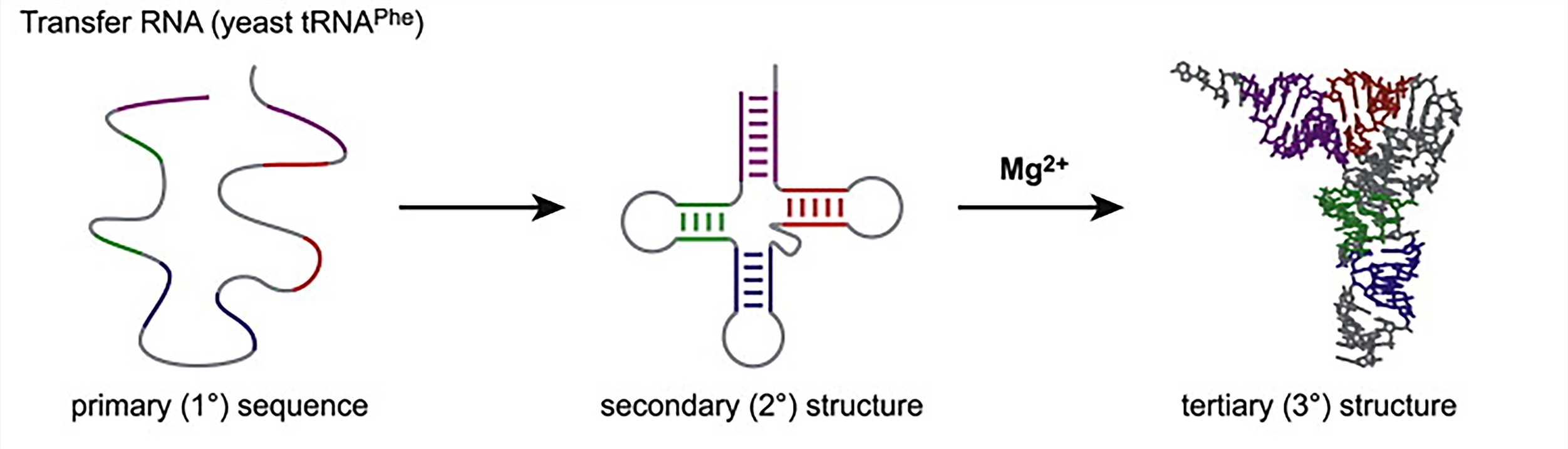 Figure 2. RNA folding hierarchy (take transfer RNA as an example)
Figure 2. RNA folding hierarchy (take transfer RNA as an example)
RNA structural motifs
Just as all three-dimensional structures of protein molecules are composed of a combination of various α- helices and β-sheets, the secondary structural motifs of RNA provide its structural building blocks. Several major RNA structural motifs are described below.
- Helices
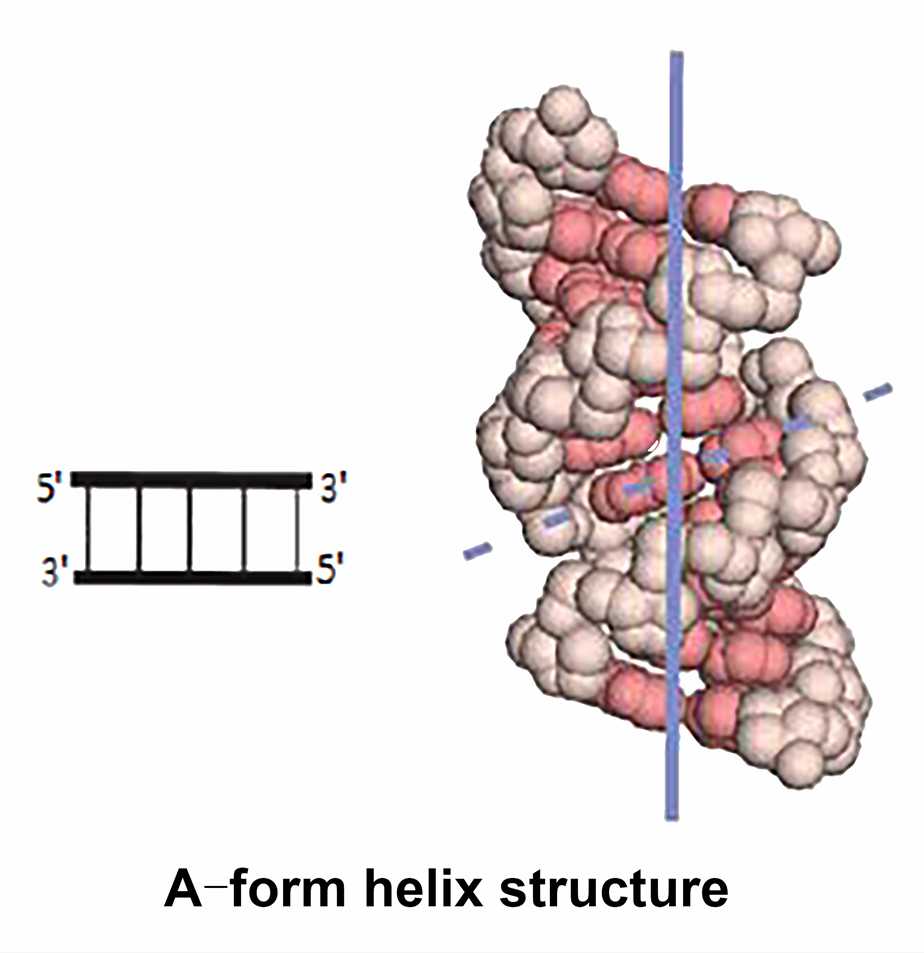
RNA often contains stretched self-complementary sequences that can be folded back to form self-complementary structures, forming an extended hairpin structure and can adopt a double helix structure. Unlike DNA, RNA cannot adopt the B-form helix because the additional 2' hydroxyl group interferes with the arrangement of the sugars in the phosphate backbone. The helical regions of RNA are often A-form helix with 11 base pairs in a circle. Watson-Crick base pairs can also be found in the double-stranded region of RNA, however, are another form of base pairing. The reason for this phenomenon is that RNA molecules do not contain perfect sequence pairing. The tendency to form a double helix by base stacking results in fewer hydrogen bonds and less ideal backbone conformation to form base pairs. For example, the guanine-uracil (G-U) base pair has only two hydrogen bonds, which is different from the normal Watson-Crick base pairing principle.
- Hairpin loops
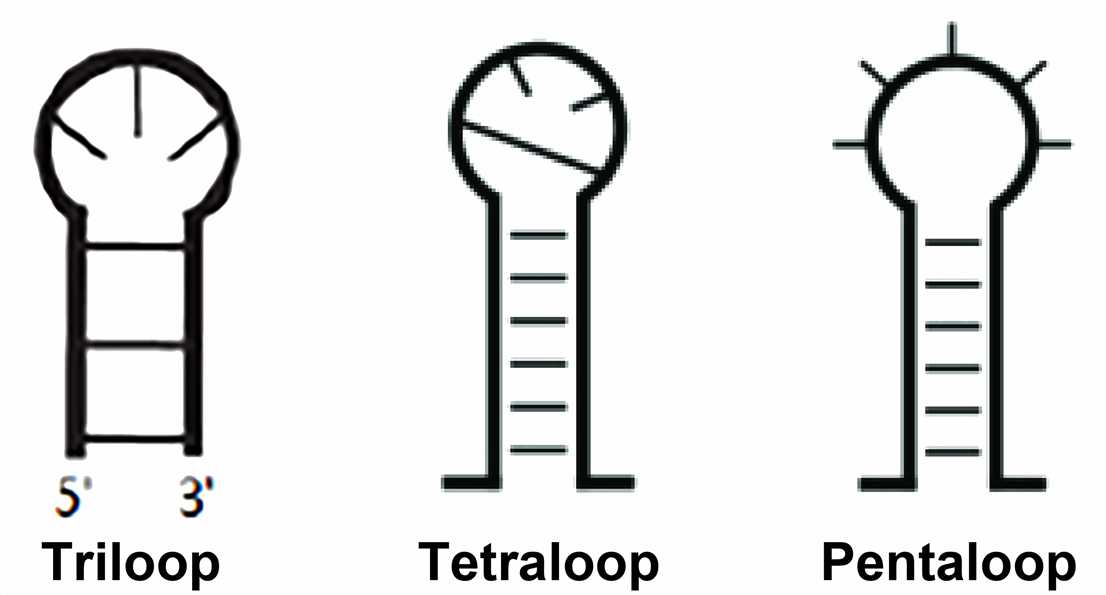
The hairpin loop is formed when the stretchable region of the RNA sequence is a completely self-complementary sequence. The hairpin loop, also called stem-loop structure, is classified according to the number of bases in the unpaired region and is known as triloop, tetraloop, pentaloop, and so on.
In the SCOR (the structural classification of RNA) database classification, structurally characterized hairpin loops must be closed in Watson-Crick pairing and vary in length from 2 to 14 ribonucleotides. The most common and studied is the tetraloop. Of these tetraloops, at least four types are characterized by their sequence and conserved structure: GNRA type, UNCG type, ANYA type, and (U/A) GNN type. In rRNA, about 70% of the tetraloops belong to either the GNRA or the UNCG family and are thermodynamically stable compared to other tetraloops. As is well-known, the GNRA is the most commonly observed tetraloop in the currently available RNA structures. Other hairpin loops include the T-loop and D-loop motifs of tRNA, lonepair triloop, and sarcin-ricin loop.
- Internal loops
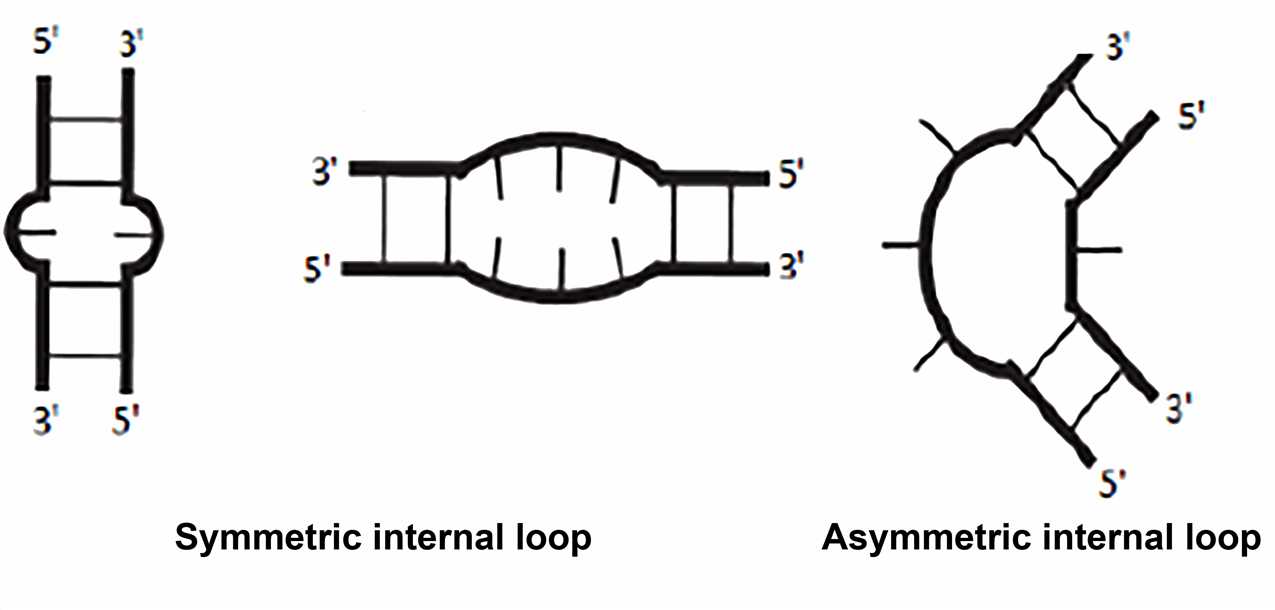
Typically, the internal loop is adjacent to two A-form helices, and the bases within the loop can be unpaired. Two types of internal loops can be distinguished: the symmetric internal loop with the same number of nucleotides inserted on both strands; the asymmetric internal loop with a different number of nucleotides inserted on the opposing strands. Sometimes insertions on only one strand are termed "bulge". Non-Watson-Crick base pairing is common in internal loops. The frequently observed motif involves extending the double helix structure by continuously forming non-canonical pairs in the symmetric internal loop. Some asymmetric internal loops have been identified and characterized as resulting in sharp turns that are important for tertiary structure formation. These include K-turn, reverse kink-turn, and hook-turn.
The internal loop is usually the binding site for the protein, such as the HIV-1 Rev protein binding to an asymmetric internal loop. An example of the internal loop that has received wide attention is the loop E motif, which was first discovered in the 1980s. Similar and conserved internal loops were found in eukaryotic 5S rRNA and PSTV viral RNA, when they were under ultraviolet light, they shared a strange portion to form a cross-combination. Subsequently, the loop E motif was also found in the sarcin-ricin loop in the 23S rRNA and was included in the binding sites of elongation factor EF-Tu and EF-G. In E. coli 5S rRNA, loop E is a known specific binding site for binding to the ribosomal protein L25. Therefore, this motif is an important active site and molecular recognition region.
- Junctions
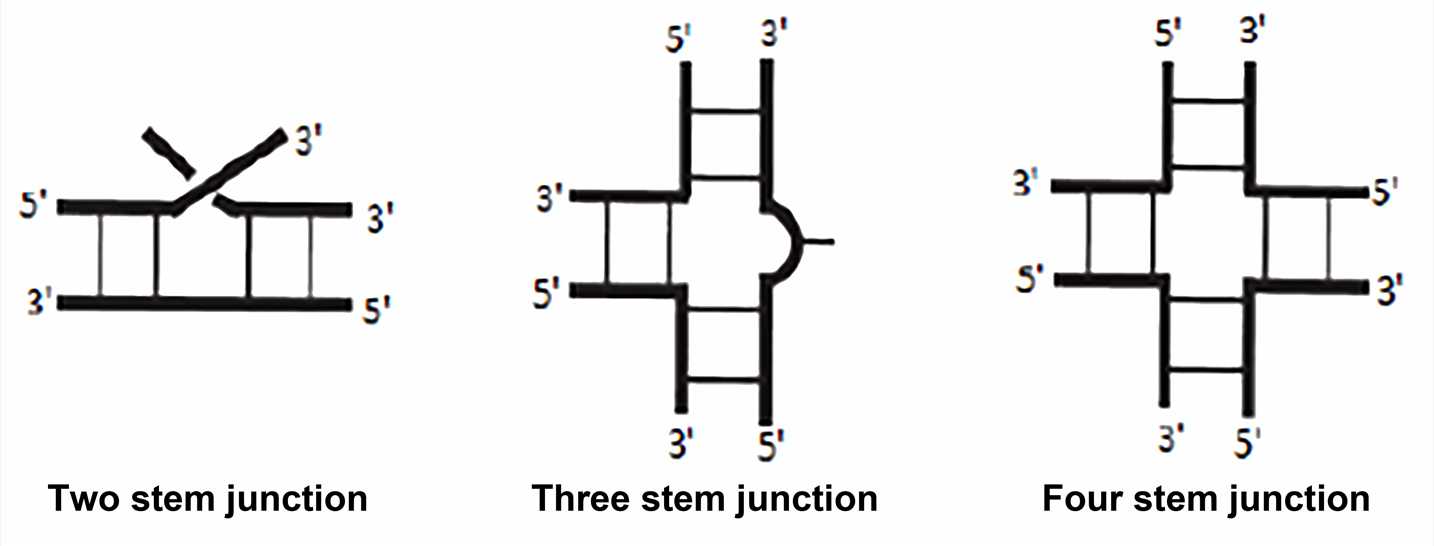
The junction is the region that joins two or more stem structures. The single strand between each stem structure may have sequences of zero or more residues, and these single strands are referred to as linking or joining regions. Although the junctions have not been as systematic or extensively studied as simpler hairpin loops and internal loops, there are some generalizations for the more common three-way and four-way junctions. Three-way junction is the most common in stable RNA, such as ribosomal RNA, viral RNA, and ribozyme. Four-way junction sometimes referred to as cruciform junction, is also a common structure, as in tRNA.
- Pseudoknots
The pseudoknot is a structure that contains two stem-loop structures in the RNA secondary structure, and the loop of the first stem-loop structure is the stem of the second stem-loop structure. The pseudoknot was first discovered in the turnip yellow mosaic virus in 1982, but until it was found in many RNA structures, most of the pseudoknots were still from the virus. The pseudoknot folds into more compact three-dimensional conformations but is not a true topological knot.
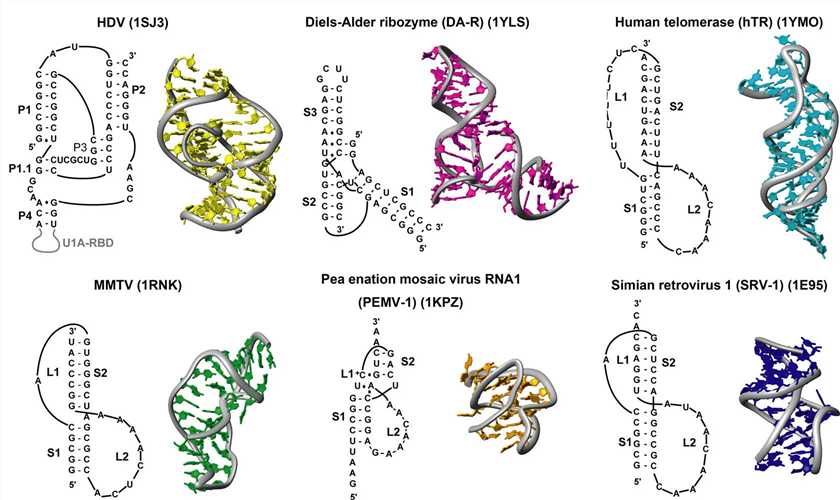 Figure 3. Sequences and structures of RNA pseudoknots
Figure 3. Sequences and structures of RNA pseudoknots
RNA three-dimensional structure and biological function
Most RNA molecules have well-defined biological functions: carrying genetic information (messenger RNA, abbreviated as mRNA, viral genome in RNA virus), forming structural entities (ribosomal RNA, abbreviated as rRNA), having recognition tasks (transfer RNA, abbreviated as tRNA and short interfering RNA, abbreviated as siRNA), catalyzing chemical reaction (ribozyme). These different roles stem from the ability of RNA molecules to take a wide range of three-dimensional structures, reaffirming the fact that RNA is more flexible than DNA. Some RNA molecules have unique conformations that perform their functions well, while others can form more flexible structures. Among them, tRNA molecules and rRNA molecules are examples of RNA ordered configurations. The coding region of mRNA is an example of a very flexible RNA molecule that can continuously pass through the ribosome, and local secondary structures need to be opened (or circumvented) during the decoding process.
To understand the role of a specific RNA molecule, we often need to know its structure. The tertiary structure of RNA can be determined by biophysical methods, such as nuclear magnetic resonance (NMR) spectroscopy, X-ray crystallography, and cryo-electron microscopy. Creative Biostructure is specialized in the field of structural biology, and we provide contract services for the structural analysis of RNA samples.
References
- Silverman S K. A forced march across an RNA folding landscape. Chemistry & Biology. 2008. 15(3): 211-213.
- Hendrix D K, et al. RNA structural motifs: building blocks of a modular biomolecule. Quarterly Reviews of Biophysics. 2005. 38(3): 221-243.
- Staple D W, Butcher S E. Pseudoknots: RNA structures with diverse functions. PLoS Biology. 2005. 3(6): e213.