Structural Research of Photosynthetic Reaction Centers
The photosynthetic reaction center (RC) is a complex of multiple proteins, pigments, and other cofactors that work together to perform the major energy transfer reactions of photosynthesis. During the reaction, the energy of photons is used to excite the electrons of the pigments, and the resulting free energy passes through a chain of nearby electron acceptors, transferring hydrogen atoms from H2O or H2S to CO2, ultimately producing glucose. In recent years, the structure of RC has been shown by various structural detection techniques, which help to reveal the reaction mechanism by which organisms convert photon energy into chemical energy.
Advances in RC structure
It is shown that all RCs have an ET structural domain consisting of two subunits each including five transmembrane helices (TMH). Among the four types of RCs, only the core of RC1 has C2 symmetry, while the other three RCs (PSII, PSI, and RC2) are structurally asymmetric. Heterodimerization is thought to be responsible for the establishment of a two-electron gate in type II RC, a protective mechanism to reduce reactive oxygen species production in PSI, and associated with water oxidation in PSII. Type I RC and PSII have six additional TMHs on each side of the ET structural domain for coordinated pigmentation.
Structural analysis of HbRC
Heliobacteria use a single RC to drive the cyclic electron transfer (ET) pathway for adenosine 5'-triphosphate (ATP) synthesis. The RC (HbRC) of heliobacteria is found to have a unique structure, which does not require a tightly bound quinone in ET. The structure shows that HbRC has an average height of about 50 Å and contains 24 TMHs, 22 from the PshA homodimer and 2 from the newly discovered PshX subunit. The ET cofactors are arranged in two identical branches around the C2 symmetry axis, and the antennae BChls form two layers in the RC, most of which are relatively close to each other.
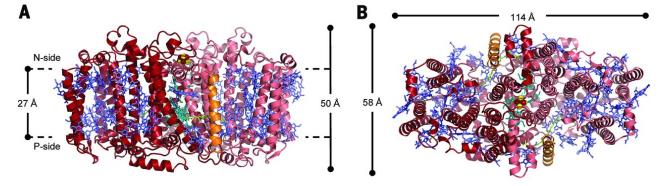 Figure 1. The overall structure of HbRC is viewed from the N-side (A) and intramembrane (B). (Gisriel C, et al., 2017)
Figure 1. The overall structure of HbRC is viewed from the N-side (A) and intramembrane (B). (Gisriel C, et al., 2017)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Photosystem I | Synechococcus elongatus | X-ray diffraction | 2.5 Å | 1JB0 |
| Trimeric photosystem I | Cyanobacterium aponinum 0216 | Cryo-EM single particle analysis | 2.7 Å | 6VPV |
| PSI monomer | Thermosynechococcus vestitus BP-1 | X-ray diffraction | 6.5 Å | 7BW2 |
| Photosystem I IsiA super-complex | Synechocystis sp. PCC 6803 | Cryo-EM single particle analysis | 3.48 Å | 6NWA |
| Photosystem I trimer | Synechocystis sp. PCC 6803 | X-ray diffraction | 2.501 Å | 5OY0 |
| Photosystem I in the presence of cytochrome c6 | Thermosynechococcus vestitus BP-1 | Cryo-EM single particle analysis | 2.85Å | 6TRA |
| Photosystem I acclimated to far-red light | Fischerella thermalis PCC 7521 | Cryo-EM single particle analysis | 3.19Å | 6PNJ |
| A red-shifted photosystem I complex | Synechocystis sp. PCC 6803 substr. Kazusa | Cryo-EM single particle analysis | 3.1 Å | 6UZV |
| GraFix PSI tetramer | Cyanophora paradoxa | Cryo-EM single particle analysis | 3.8 Å | 7DR2 |
| Photosystem I-LHCI-Lhca5 supercomplex | Hordeum vulgare subsp. spontaneum | Cryo-EM single particle analysis | 3.4 Å | 7EW6 |
| Photosynthetic oxygen evolving center | Thermosynechococcus vestitus | X-ray diffraction | 3.5 Å | 1S5L |
| Photosystem II, first illuminated state | Thermosynechococcus vestitus | X-ray diffraction | 5.9 Å | 4IXR |
| PSII-I prime (PSII with Psb28, and Psb34) | Thermosynechococcus vestitus BP-1 | Cryo-EM single particle analysis | 2.68 Å | 7NHQ |
| PSII-I (PSII with Psb27, Psb28, and Psb34) | Thermosynechococcus vestitus BP-1 | Cryo-EM single particle analysis | 2.72 Å | 7NHP |
| PSII-M | Thermosynechococcus vestitus BP-1 | Cryo-EM single particle analysis | 2.66 Å | 7NHO |
| Photosystem II | Thermosynechococcus vestitus | X-ray diffraction | 3 Å | 2AXT |
| Monomeric photosystem II | Thermostichus vulcanus | Cryo-EM single particle analysis | 2.78 Å | 7EDA |
| PSII intermediate Psb28-PSII complex | Thermostichus vulcanus | Cryo-EM single particle analysis | 3.14 Å | 7DXH |
| Photosystem II complex | Thermostichus vulcanus | X-ray diffraction | 1.9 Å | 3WU2 |
| Photosystem II structure in the S1 state | Thermostichus vulcanus | X-ray diffraction | 2.35 Å | 7CJI |
| Photosystem II structure in the S2 state | Thermostichus vulcanus | X-ray diffraction | 2.4 Å | 7CJJ |
| Homodimeric reaction center | Heliomicrobium modesticaldum | X-ray diffraction | 2.2 Å | 5V8K |
| The whole photosynthetic complex | Chlorobaculum tepidum TLS | Cryo-EM single particle analysis | 2.5 Å | 7Z6Q |
| Photosynthetic reaction center | Thermochromatium tepidum | X-ray diffraction | 2.2 Å | 1EYS |
| Photosynthetic reaction center (ubiquinone-2 complex) | Blastochloris viridis | X-ray diffraction | 2.45 Å | 2PRC |
Table 1. Structural research of photosynthetic reaction centers.
Cryo-electron microscopy (cryo-EM) and X-ray crystallography are commonly used to determine the structure of photosynthetic reaction centers. Both techniques provide high-resolution images of protein structures and are helpful in defining conserved patterns of cofactors and core proteins in the reaction centers of diverse organisms.
Creative Biostructure is devoted to providing excellent services on 3D structural studies of membrane proteins. We utilize cutting-edge testing equipment and methods to provide our clients with the best service and results. Our extensively trained and certified laboratory staff has the expertise and proficiency to ensure accurate and reliable data delivery. If you are interested in our services, please contact us for more details.
References
- Gisriel C, et al. Structure of a symmetric photosynthetic reaction center–photosystem. Science. 2017. 357(6355):1021-1025.
- Gisriel CJ, et al. Recent advances in the structural diversity of reaction centers. Photosynth Res. 2021. 149(3):329-343.
- Chen JH, et al. Architecture of the photosynthetic complex from a green sulfur bacterium. Science. 2020. 370(6519): eabb6350.