Structural Research of Protein O-mannosyltransferases (POMT/PMT)
Protein O-mannosyltransferases (POMT/PMT) catalyze the addition of mannose to serine or threonine residues of secreted proteins. It depends on specific isoforms of the three Pmt1, 2, and 4 subfamilies. In mammals and insects, PMTs are essential for cell differentiation and development. In fungi, PMT determines cell wall structure and integrity, as well as cell differentiation and virulence. Although the necessity of Pmts for fungal pathogenesis has been well established in several pathogens, structural investigations of Pmts remain a hot topic.
Structural analysis of Pmts
Research has shown that Pmt is a multi-span endoplasmic reticulum (ER) membrane protein containing three structural domains. The two ER transmembrane α-helical regions are PMT and 4TMC. The third structural domain is the central MIR structural domain, which is shared by mannosyltransferase, inositol trisphosphate receptor, and lanylphosphatidylcholine receptor, and has a β-trefoil fold. The MIR structural domain is located in the ER lumen, and it interacts directly with the substrate, conferring target specificity to Pmt. In addition, Pmt1 is characterized by a long C-terminal region containing a disordered structural domain that is conserved in most fungal Pmt1 proteins.
Overall structural analysis of Pmt1-Pmt2 complexes
Pmts usually act by forming dimers. In yeast, Pmt1 forms a complex with Pmt2. The researchers used cryo-electron microscopy to determine establish and refine an atomic model of the complete complex. The Pmt1-Pmt2 complex has pseudo-double symmetry. both Pmt1 and Pmt2 contain transmembrane regions and a luminal MIR structural domain. the MIR structural domain contains three MIR motifs that form β-trefoils, each containing four β-strands and a short α-helix. the transmembrane regions of Pmt1 and Pmt2 are connected by contacts located in the cytoplasmic and luminal regions, leading to a large rhombic shape in the center. resulting in a large rhombic cavity in the center. There are five potential N-glycosylation sites in the complex, three (N390, N513, and N743) in Pmt1 and two (N131 and N403) in Pmt2.
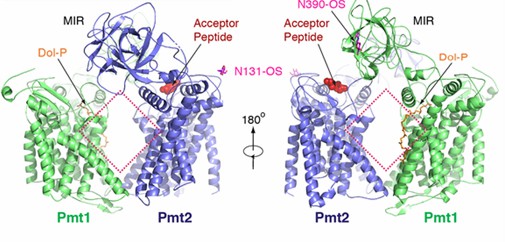 Figure 1. Schematic representation of the overall structure of the Pmt1-Pmt2 complex. (Bai L, et al., 2019)
Figure 1. Schematic representation of the overall structure of the Pmt1-Pmt2 complex. (Bai L, et al., 2019)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Protein O-mannosyltransferase Pmt1-Pmt2 complex bound to the sugar donor | Saccharomyces cerevisiae W303 | Cryo-EM single particle analysis | 3.2 Å | 6P2R |
| Protein O-mannosyltransferase Pmt1-Pmt2 complex bound to the sugar donor and a peptide acceptor | Saccharomyces cerevisiae W303 | Cryo-EM single particle analysis | 3.2 Å | 6P25 |
| MIR domain (aa 337-532) of Pmt2 | Saccharomyces cerevisiae | X-ray diffraction | 1.35 Å | 6P28 |
| Pmt2-MIR domain with bound ligands | Saccharomyces cerevisiae | X-ray diffraction | 1.6 Å | 6ZQP |
| Pmt3-MIR domain with bound ligands | Saccharomyces cerevisiae | X-ray diffraction | 1.9 Å | 6ZQQ |
| Alpha-1,3-mannosyltransferase MNT2, Mn/GDP-mannose form | Saccharomyces cerevisiae W303 | X-ray diffraction | 2.8 Å | 7XJV |
| Alpha1,2-mannosyltransferase Kre2p/Mnt1p | Saccharomyces cerevisiae | X-ray diffraction | 2.01 Å | 1S4N |
| The mannosyltransferase Ktr4p | Saccharomyces cerevisiae S288C | X-ray diffraction | 2.21 Å | 5A08 |
| The mannosyltransferase Ktr4p in complex with GDP | Saccharomyces cerevisiae S288C | X-ray diffraction | 1.9 Å | 5A07 |
| Mannosyltransferase PcManGT | Pyrobaculum calidifontis JCM 11548 | X-ray diffraction | 2.7 Å | 6YV7 |
Table 1. Structural research of protein O-mannosyltransferases (POMT/PMT).
Creative Biostructure provides cutting-edge structural analysis services, such as X-ray crystallography, cryo-electron microscopy (cryo-EM), and nuclear magnetic resonance (NMR). With our team of highly skilled experts and cutting-edge equipment, we can reveal the complex structures of biomolecules and their complexes. With cutting-edge technologies and methodologies, we can help clients gain insights into the structure and function of protein O-mannosyltransferases (POMT/PMT), thereby helping to develop novel therapeutic strategies for related diseases. If you have any questions, please contact us and we will use our expertise to unlock the secrets of structural biology.
References
- Bai L, et al. Structure of the eukaryotic protein O-mannosyltransferase Pmt1-Pmt2 complex. Nat Struct Mol Biol. 2019.26(8):704-711.
- Pejenaute-Ochoa MD, et al. Structural, Evolutionary, and Functional Analysis of the Protein O-Mannosyltransferase Family in Pathogenic Fungi. J Fungi (Basel). 2021.7(5):328.
- Lengeler, K. B., et al. Protein-O-mannosyltransferases in virulence and development.Cellular and Molecular Life Sciences. 2007.65(4), 528–544.