Structural Research of SWEET and semiSWEET Transporters
SWEET transporters were first identified in Arabidopsis thaliana with the aid of a fluorescent biosensor and validated as unique sugar transport proteins. Their homologues are present in plants, animals, protozoa, and bacteria which catalyze the facilitated diffusion of sugars across the plasma membrane or endoplasmic reticulum membrane. Research has revealed that eukaryotic SWEET family proteins have seven transmembrane segments (TMS) that produce a similar conformation to the bacterial SemiSWEET dimer in a “3+1+3” repeating arrangement. Prokaryotic semiSWEET is small and contains parallel homodimers of triple helix bundles (THBs) approximately 100 amino acids long.
Advances in research on the structure of semiSWEET in bacteria
Many bacterial homologues of SWEET proteins have only 3 TMS and are half the size. Like most eukaryotic proteins in the family, other bacterial homologues have seven TMS. bacterial semiSWEET consists of a triple helix bundle in a 1-3-2 conformation, with TM3 sandwiched between TM1 and TM2. The structure also shows tryptophan and asparagine residues interacting with sugars, and point mutations of these residues to alanine disrupt the hexose transport function of semiSWEET.
Structural analysis of the plant SWEET protein
In plants, SWEET proteins use intra- and extracellular sugar concentration gradients to transport them across membranes, rather than relying on proton gradients. Plant SWEET proteins are members of the MtN3/salivary family, which typically contains seven TMs, with the N-terminus and C-terminus located in the outer and inner cytoplasm, respectively. The TM4 divides SWEET proteins into two MtN3/salivary structural domains, each of which contains three TMs, forming a "3-1-3" structure. The three TMs of each MtN3/salivary domain are arranged in the form of "TM1-TM3-TM2" to form a triple helix bundle (THB).
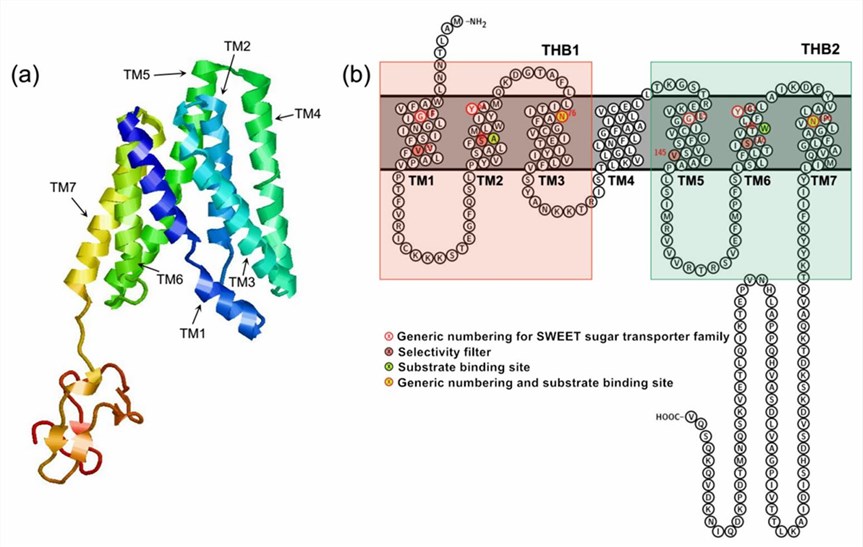 Figure 1. Structural features of the plant SWEET transporter. (Ji J, et al., 2022)
Figure 1. Structural features of the plant SWEET transporter. (Ji J, et al., 2022)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Eukaryotic SWEET transporter | Oryza sativa Japonica Group | X-ray diffraction | 3.103 Å | 5CTG |
| Eukaryotic SWEET transporter | Oryza sativa Japonica Group | X-ray diffraction | 3.69 Å | 5CTH |
| SemiSWEET | Vibrio sp. N418 | X-ray diffraction | 1.698 Å | 4QND |
| SemiSWEET in the inward-open conformation | Escherichia coli UMEA 3162-1 | X-ray diffraction | 2 Å | 4X5M |
| SemiSWEET in the inward-open and outward-open conformations | Escherichia coli UMEA 3162-1 | X-ray diffraction | 3 Å | 4X5N |
| SemiSWEET in an occluded state | Leptospira biflexa serovar Patoc strain 'Patoc 1 (Paris) | X-ray diffraction | 2.388 Å | 4QNC |
| SemiSWEET Q20A mutant | Leptospira biflexa serovar Patoc strain 'Patoc 1 (Paris) | X-ray diffraction | 2.78 Å | 5UHQ |
| SemiSWEET D57A mutant | Leptospira biflexa serovar Patoc strain 'Patoc 1 (Paris) | X-ray diffraction | 2.8 Å | 5UHS |
| Bacterial homologue of SWEET transporters | Thermodesulfovibrio yellowstonii DSM 11347 | X-ray diffraction | 2.4 Å | 4RNG |
| Sugar transporter of AtSWEET13 in inward-facing state with a substrate analog | Arabidopsis thaliana | X-ray diffraction | 2.788 Å | 5XPD |
Table 1. Structural research of the SWEET and semiSWEET transporters.
Creative Biostructure offers a comprehensive structural analysis services for SWEET and semiSWEET transporters. Our scientists use cutting-edge equipment and techniques including X-ray crystallography, cryo-electron microscopy (cryo-EM), and NMR spectroscopy to determine the three-dimensional structure of proteins and other biomolecules. In addition, we support the production, purification, and characterization of proteins to ensure the best possible results for your project.
As an industry-leading provider of structural analysis services, we specialize in the elucidation of complex molecular structures and can offer our clients unparalleled structural analysis services. We welcome all researchers and organizations to contact us for advice and consultation. Please contact us now to find out how we can help advance your research.
References
- Ji J, et al. Plant SWEET family of sugar transporters: structure, evolution and biological functions. Biomolecules. 2022. 12(2): 205.
- Feng L, Frommer WB. Structure and function of SemiSWEET and SWEET sugar transporters. Trends Biochem Sci. 2015. 40(8): 480-486.
- Wang J, et al. Crystal structure of a bacterial homologue of SWEET transporters. Cell Res. 2014. 24(12): 1486-1489.