Structural Research of Virus Coat Proteins
The coat/capsid is the protein shell of a virus and consists of several proteins comprising oligomeric (repetitive) structural subunits. The primary function of viral coat/capsid protein (CP) is to form capsids that protect the viral genome from degradation. In addition to structural functions, CPs have essential activities in the viral infection cycle and the host defense response to viral infection. In recent years, a major goal of virus studies has been how many of the viral lineages can be identified from structural studies of viral CPs.
Progress in research on the major CPs of viruses
The structures of the major CPs of several viruses have been reported, such as the four-helix bundle of ssRNA tobacco mosaic virus (TMV), the α/β-folded structure of the Leviviridae bacteriophage virus, and the similar three-domain glycoproteins of alphaviruses and flaviviruses. In addition, two ancient lineages of DNA viruses have been identified based on the CPs and vesicle structures of dsDNA icosahedral viruses. The structure of the first virus, characterized by a jelly-like fold (single or double layer), is first found in the pod membranes of adenoviruses and phage PRD1. The second type of virus is found in bacterial capsid viruses, herpesviruses, as well as in archaea and bacterial capsid proteins.
Structural analysis of potato Y virus (PVY) CP
PVY is one of the most economically essential plant pathogens. In recent years, researchers have used cryo-electron microscopy to determine the near-atomic structure of the PVY curved filamentous virion by a powerful high-resolution analytical method, revealing luminal interactions between the extended carboxyl-terminal region of the capsid protein unit and the viral RNA. The structure shows that the PVY virus particle consists of an arrangement of CP left-handed helices assembled around the viral ssRNA. The spherical core subdomain contains seven α-helices and a β-hairpin, and the helically arranged core assembles into a protein layer that protects the viral ssRNA from environmental influences.
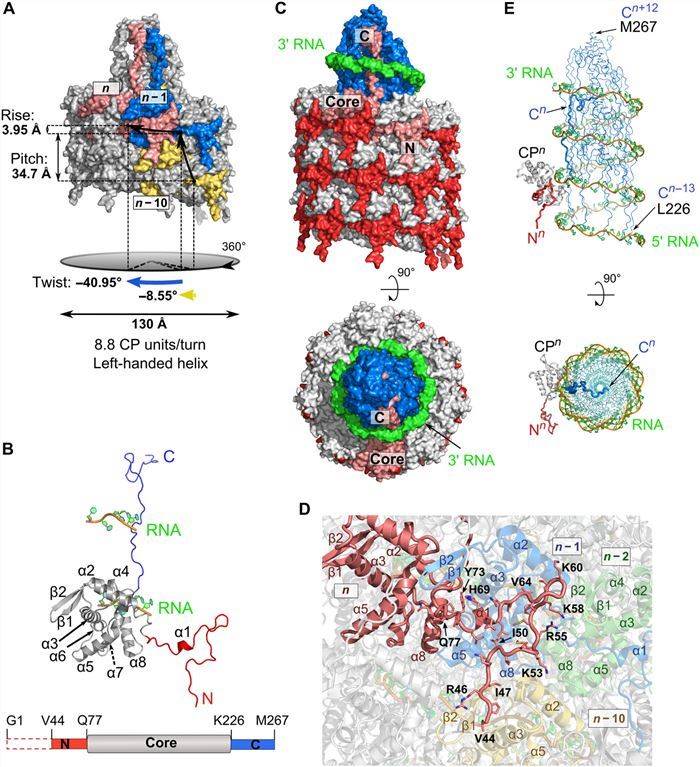 Figure 1. Structural features of PVY coat protein. (Kežar A, et al., 2019)
Figure 1. Structural features of PVY coat protein. (Kežar A, et al., 2019)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Rice yellow mottle virus | Rice yellow mottle virus | X-ray diffraction | 2.8 Å | 1F2N |
| Norwalk virus capsid | Norwalk virus | X-ray diffraction | 3.4 Å | 1IHM |
| Tomato aspermy virus | Tomato aspermy virus | X-ray diffraction | 3.4 Å | 1LAJ |
| Bacteriophage alpha3 assembly | Escherichia phage alpha3 | X-ray diffraction | 3.5 Å | 1M06 |
| The phiX174 DNA binding protein J in two different capsid environments. | Escherichia phage alpha3 | X-ray diffraction | 3.5 Å | 1RB8 |
| MS2-RNA hairpin (2thiouracil-5) complex | Escherichia phage MS2 | X-ray diffraction | 2.68 Å | 2C4Y |
| MS2-RNA hairpin (C-10) complex | Escherichia phage MS2 | X-ray diffraction | 2.7 Å | 2IZM |
| A native calicivirus | San Miguel sea lion virus 4 | X-ray diffraction | 3.2 Å | 2GH8 |
| P Domain of Norovirus VA387 | Norovirus | X-ray diffraction | 2.2 Å | 2OBR |
| Adeno-associated virus serotype 8 | Adeno-associated virus - 8 | X-ray diffraction | 2.6 Å | 2QA0 |
| The picobirnavirus capsid | Rabbit picobirnavirus | X-ray diffraction | 3.4 Å | 2VF1 |
| Barmah Forest virus structural proteins | Barmah Forest virus | Cryo-EM single particle analysis | 5 Å | 2YEW |
| Hepatitis E virus ORF2 (Genotype 3) | Paslahepevirus balayani | X-ray diffraction | 3.6 Å | 2ZTN |
| Backbone trace of the capsid protein dimer of a fungal partitivirus | Penicillium stoloniferum virus S | Cryo-EM single particle analysis | 4.7 Å | 3IYM |
| Penicillium chrysogenum virus (PcV) capsid | Penicillium chrysogenum virus (isolate Caston) | Cryo-EM single particle analysis | 4.1 Å | 3J3I |
| Feline calicivirus capsid protein | Feline calicivirus | X-ray diffraction | 3.4 Å | 3M8L |
| Penaeus stylirostris densovirus capsid | Decapod penstyldensovirus 1 | X-ray diffraction | 2.5 Å | 3N7X |
| Triatoma virus (TrV) | Triatoma virus | X-ray diffraction | 2.5 Å | 3NAP |
| AAV8 capsid transitions associated with endosomal trafficking | Adeno-associated virus - 8 | X-ray diffraction | 2.7 Å | 3RA2 |
| Capsid protein (110-267) from Aura virus | Aura virus | X-ray diffraction | 1.81 Å | 4AGK |
| Hexameric HBc149 Y132A | Hepatitis B virus | X-ray diffraction | 3 Å | 4BMG |
| Rubella virus capsid protein (residues 127-277) | Rubella virus strain M33 | X-ray diffraction | 2.663 Å | 4HAR |
| Cucumber necrosis virus | Cucumber necrosis virus | X-ray diffraction | 2.8891 Å | 4LLF |
| Orsay virus-like particle | Orsay virus | X-ray diffraction | 3.25 Å | 4NWV |
| Norovirus OIF P domain | Norovirus NLV/IF1998/2003/Iraq | X-ray diffraction | 1.19 Å | 4RLZ |
Table 1. Structural research of the virus coat proteins.
At Creative Biostructure, we pride ourselves on our cutting-edge structural analysis services. With years of experience in the field, our team of expert researchers and state-of-the-art technologies enable us to conduct comprehensive studies on the structure of virus coat proteins.
Our team of specialists has the experience and cutting-edge equipment to fully support your research. Our experts are committed to advancing science by utilizing advanced techniques such as X-ray crystallography, cryo-electron microscopy (cryo-EM), and NMR spectroscopy to obtain high-resolution structural data. If you are interested in protein structure analysis and related research areas, please feel free to contact us for more details.
References
- Kežar A, et al. Structural basis for the multitasking nature of the potato virus Y coat protein. Sci Adv. 2019. 5(7): eaaw3808.
- Kavčič L, et al. From structural polymorphism to structural metamorphosis of the coat protein of flexuous filamentous potato virus Y. Commun Chem. 2024. 7(1): 14.
- Goulet A, et al. Acidianus filamentous virus 1 coat proteins display a helical fold spanning the filamentous archaeal viruses lineage. Proc Natl Acad Sci U S A. 2009. 106(50): 21155-21160.
