Structural Research of Monomeric/Dimeric Beta-Barrel Membrane Proteins
Beta-barrel membrane proteins are a class of proteins that are embedded in the lipid bilayer of the cell membrane and play a vital role in the transport of various molecules across the membrane. These proteins consist of multiple β-strands that form a cylindrical shape with a central pore and can exist as monomers or form dimers or higher-order oligomers in the membrane.
Recent advances in structural biology techniques have enabled the determination of the high-resolution structures of several beta-barrel membrane proteins in their monomeric and dimeric forms. For example, the dynamic NMR structures of AlkL, a protein that plays a role in transporting hydrophobic molecules across the membrane, revealed a β-barrel structure with hydrophobic residues lining the inside of the barrel. The crystal structure of OmpF, a protein that forms a dimeric structure in the membrane, showed that the dimerization is mediated by hydrogen bonds and van der Waals interactions between the β-strands.
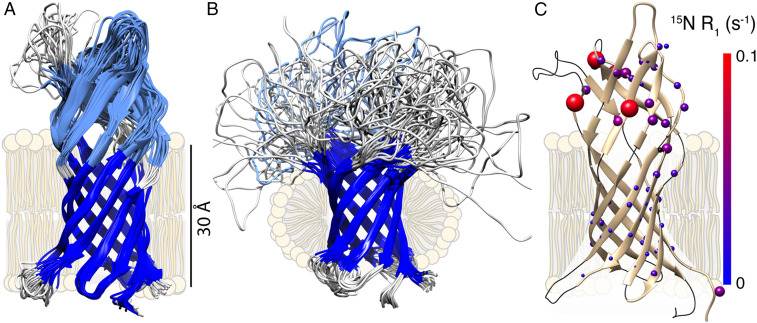 Figure 1. NMR structure and dynamics of AlkL. (Schubeis T, et al., 2020)
Figure 1. NMR structure and dynamics of AlkL. (Schubeis T, et al., 2020)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| TolC outer membrane protein (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.10 Å | 1EK9 |
| TolC outer membrane protein, ligand blocked | Escherichia coli | X-ray diffraction | 2.75 Å | 1TQQ |
| TolC outer membrane protein (Y362F, R367E), partially open state (expressed in E. coli) | Escherichia coli | X-ray diffraction | 3.20 Å | 2VDE |
| TolC outer membrane protein in a nanodisc with colicin E1 fragment (expressed in E. coli) | Escherichia coli | Cryo-EM single particle analysis | 3.09 Å | 6WXH |
| TolC outer membrane protein in complex with KlebC (expressed in E. coli) | Klebsiella quasipneumoniae | Cryo-EM single particle analysis | 3.20 Å | 7NG8 |
| CmeC bacterial multi-drug efflux transporter outer membrane channel (expressed in E. coli) | Campylobacter jejuni | X-ray diffraction | 2.37 Å | 4MT4 |
| VceC outer membrane protein (expressed in E. coli) | Vibrio cholerae | X-ray diffraction | 1.80 Å | 1YC9 |
| OprM drug discharge outer membrane protein (expressed in Pseudomonas aeruginosa) | Pseudomonas aeruginosa | X-ray diffraction | 2.56 Å | 1WP1 |
| OprM drug discharge outer membrane protein (expressed in E. coli) | Pseudomonas aeruginosa | X-ray diffraction | 2.40 Å | 3D5K |
| OprN drug discharge outer membrane protein, I4 space group (expressed in E. coli) | Pseudomonas aeruginosa | X-ray diffraction | 2.70 Å | 5AZO |
| OprJ drug discharge outer membrane protein (expressed in E. coli) | Pseudomonas aeruginosa | X-ray diffraction | 3.10 Å | 5AZS |
| ST50 discharge outer membrane protein (expressed in E. coli) | Salmonella enterica | X-ray diffraction | 2.98 Å | 5BUN |
| CusC heavy metal discharge outer membrane protein | Escherichia coli | X-ray diffraction | 2.30 Å | 3PIK |
| CusC heavy metal discharge outer membrane protein (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.09 Å | 4K7R |
| apo BtuB cobalamin transporter (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.00 Å | 1NQE |
| BtuB with bound colicin E3 R-domain (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.75 Å | 1UJW |
| apo BtuB by in meso crystallization (expressed in E. coli) | Escherichia coli | X-ray diffraction | 1.95 Å | 2GUF |
| BtuB in complex with TonB (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.10 Å | 2GSK |
| BtuB with bound colicin E2 R-domain (expressed in E. coli) | Escherichia coli | X-ray diffraction | 3.50 Å | 2YSU |
| apo BtuB V10R1 spin-labeled (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.44 Å | 3M8B |
| Colicin I receptor Cir in complex with Colicin Ia binding domain (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.50 Å | 2HDI |
| OmpA (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.50 Å | 1BXW |
| OmpA (expressed in E. coli) | Escherichia coli | X-ray diffraction | 1.65 Å | 1QJP |
| OmpA (expressed in E. coli) | Escherichia coli | Solution NMR | / | 1G90 |
| OmpA (expressed in E. coli) | Escherichia coli | Solution NMR | / | 2GE4 |
| OmpA with four shortened loops (expressed in E. coli) | Escherichia coli | Solution NMR | / | 2JMM |
| OmpA (expressed in E. coli) | Klebsiella pneumoniae | Solution NMR | / | 2K0L |
| OmpT outer membrane protease (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.60 Å | 1I78 |
| Pla Plasminogen activator (native 1) (expressed in E. coli) | Yersinia pestis | X-ray diffraction | 1.85 Å | 2X55 |
| OmpW outer membrane protein (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.70 Å | 2F1V |
| OmpW outer membrane protein (expressed in E. coli) | Escherichia coli | Solution NMR | / | 2MHL |
| AlkL passive importer of hydrophobic molecules in DMPC lipid bilayer (expressed in E. coli) | Pseudomonas oleovorans | Solid-state NMR | / | 6QWR |
| CarO outer membrane protein, isoform 1 (expressed in E. coli) | Acinetobacter baumannii | X-ray diffraction | 2.70 Å | 4RL9 |
| OprF outer membrane protein, N-terminal β-barrel domain (expressed in E. coli) | Pseudomonas aeruginosa | X-ray diffraction | 1.60 Å | 4RLC |
| OprG outer membrane protein (expressed in E. coli) | Pseudomonas aeruginosa | X-ray diffraction | 2.40 Å | 2X27 |
| OprH, outer membrane protein H (expressed in E. coli) | Pseudomonas aeruginosa | Solution NMR | / | 2LHF |
| OmpX (expressed in E. coli) | Escherichia coli | X-ray diffraction | 1.90 Å | 1QJ8 |
| OmpX (expressed in E. coli) | Escherichia coli | Solution NMR | / | 1ORM |
| OmpX (expressed in E. coli) | Escherichia coli | Solution NMR | / | 1Q9F |
| OmpX in optimized nanodiscs (expressed in E. coli) | Escherichia coli | Solution NMR | / | 2M06 |
| Ail adhesion protein (expressed in E. coli) | Yersinia pestis | X-ray diffraction | 1.80 Å | 3QRA |
| Ail adhesion protein, decylphosphocholine micelles (expressed in E. coli) | Yersinia pestis | Solution NMR | / | 2N2M |
| Ail adhesion protein (expressed in E. coli) | Yersinia pestis | Solution NMR | / | 5VJ8 |
| TtoA Outer Membrane Protein (OMP) (expressed in Thermus thermophilus) | Thermus thermophilus | X-ray diffraction | 2.80 Å | 3DZM |
| OmpLA (PldA) outer membrane phospholipase A monomer (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.17 Å | 1QD5 |
| OmpLA (PldA) outer membrane phospholipase A monomer with Ca++ (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.60 Å | 1FW2 |
| OmpLA (PldA) active-site mutant (N156A), pH 6.1 (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.50 Å | 1ILZ |
| OpcA adhesin protein | Neisseria meningitidis | X-ray diffraction | 2.03 Å | 1K24 |
| OpcA adhesin protein (expressed in E. coli) | Neisseria meningitidis | X-ray diffraction | 1.95 Å | 2VDF |
| NspA surface protein (expressed in E. coli) | Neisseria meningitidis | X-ray diffraction | 2.55 Å | 1P4T |
| NanC Porin, model for KdgM porin family (expressed in E. coli) | Escherichia coli | X-ray diffraction | 1.80 Å | 2WJR |
| PagL LPS 3-O-deacylase (expressed in E. coli) | Pseudomonas aeruginosa | X-ray diffraction | 2.00 Å | 2ERV |
| LpxR lipid A deacylase (expressed in E. coli) | Salmonella enterica | X-ray diffraction | 1.90 Å | 3FID |
| PagP outer membrane palimitoyl transferease (expressed in E. coli) | Escherichia coli | Solution NMR | / | 1MM4 |
| PagP outer membrane palimitoyl transferease (expressed in E. coli) | Escherichia coli | X-ray diffraction | 1.90 Å | 1THQ |
| PagP outer membrane palimitoyl transferease crystallized from SDS/Co-solvent (expressed in E. coli) | Escherichia coli | X-ray diffraction | 1.40 Å | 3GP6 |
| FadL long-chain fatty acid transporter (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.60 Å | 1T16 |
| FadL long-chain fatty acid transporter A77E/S100R mutant (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.50 Å | 3DWN |
| FadL long-chain fatty acid transporter D348R mutant (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.60 Å | 3PGR |
| FadL homologue long-chain fatty acid transporter (expressed in E. coli) | Pseudomonas aeruginosa | X-ray diffraction | 2.20 Å | 3DWO |
| YebT lipid transporter, domains 1-4 (expressed in E. coli) | Escherichia coli | Cryo-EM single particle analysis | 3.10 Å | 6KZ3 |
| FauA alcaligin outer membrane transporter (expressed in E. coli) | Bordetella pertussis | X-ray diffraction | 2.33 Å | 3EFM |
| TodX hydrocarbon transporter (expressed in E. coli) | Pseudomonas putida | X-ray diffraction | 2.60 Å | 3BS0 |
| TbuX hydrocarbon transporter (expressed in E. coli) | Ralstonia pickettii | X-ray diffraction | 3.20 Å | 3BRY |
| Tsx nucleoside transporter (apoprotein) (expressed in E. coli) | Escherichia coli | X-ray diffraction | 3.01 Å | 1TLY |
| FhuA, Ferrichrome-iron receptor without ligand | Escherichia coli | X-ray diffraction | 2.74 Å | 1BY3 |
| FhuA (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.50 Å | 2FCP |
| FhuA-AW140-LPS (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.50 Å | 1QFG |
| FhuA in complex with albomycin (expressed in E. coli) | Escherichia coli | X-ray diffraction | 3.10 Å | 1QKC |
| FhuA in complex with lipopolysaccharide and rifamycin CGP4832 (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.90 Å | 1FI1 |
| FhuA in complex withTonB (expressed in E. coli) | Escherichia coli | X-ray diffraction | 3.30 Å | 2GRX |
| FhuA in complex with lasso peptide microcin J25 (MccJ25) (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.30 Å | 4CU4 |
| FhuA in complex with the superinfection exclusion lipoprotein Llp (expressed in E. coli) | Escherichia coli | X-ray diffraction | 3.37 Å | 8A60 |
| FyuA siderophore transporter (expressed in E. coli) | Yersinia pestis | X-ray diffraction | 3.20 Å | 4EPA |
| FepA, Ferric enterobactin receptor | Escherichia coli | X-ray diffraction | 2.40 Å | 1FEP |
| Fiu Ferric-Catecholate import receptor, open C222(1) crystal form (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.10 Å | 6BPN |
| FecA, siderophore transporter (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.00 Å | 1KMO |
| FecA, siderophore transporter (no ligand) (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.50 Å | 1PNZ |
| FecA, siderophore transporter periplasmic signalling domain (expressed in E. coli) | Escherichia coli | Solution NMR | / | 2D1U |
| PiuA siderophore receptor (expressed in E. coli) | Pseudomonas aeruginosa | X-ray diffraction | 1.90 Å | 5FOK |
| PiuD siderophore receptor (expressed in E. coli) | Pseudomonas aeruginosa | X-ray diffraction | 2.30 Å | 5NEC |
| PiuA siderophore receptor (expressed in E. coli) | Acinetobacter baumannii | X-ray diffraction | 1.94 Å | 5FP1 |
| PirA siderophore receptor (expressed in E. coli) | Pseudomonas aeruginosa | X-ray diffraction | 2.97 Å | 5FP2 |
| PirA siderophore receptor (expressed in E. coli) | Acinetobacter baumannii | X-ray diffraction | 2.83 Å | 5FR8 |
| HasR heme-uptake receptor in complex with HasA hemophore and heme (expressed in E. coli) | Serratia marcescens | X-ray diffraction | 2.70 Å | 3CSL |
| ShuA heme-uptake receptor in complex with HasA hemophore and heme (expressed in E. coli) | Shigella dysenteriae | X-ray diffraction | 2.60 Å | 3FHH |
| FptA pyochelin siderophore transporter | Pseudomonas aeruginosa | X-ray diffraction | 2.00 Å | 1XKW |
| FptA pyochelin siderophore transporter (expressed in Pseudomonas fluorescens) | Pseudomonas fluorescens | X-ray diffraction | 3.26 Å | 3QLB |
| FpvA, Pyoverdine receptor (expressed in Pseudomonas aeruginosa) | Pseudomonas aeruginosa | X-ray diffraction | 3.60 Å | 1XKH |
| FpvA, Pyoverdine receptor (apo form) (expressed in Pseudomonas aeruginosa) | Pseudomonas aeruginosa | X-ray diffraction | 2.77 Å | 2O5P |
| FpvA, Full-length structure bound to iron-pyoverdine (expressed in Pseudomonas aeruginosa) | Pseudomonas aeruginosa | X-ray diffraction | 2.73 Å | 2IAH |
| AlgE alginate export protein (expressed in E. coli) | Pseudomonas aeruginosa | X-ray diffraction | 2.30 Å | 3RBH |
| AlgE alginate export protein (expressed in E. coli) | Pseudomonas aeruginosa | X-ray diffraction | 1.90 Å | 4AFK |
| AlgE alginate export protein at 100 K (expressed in E. coli) | Pseudomonas aeruginosa | X-ray diffraction | 2.90 Å | 4XNL |
| AlgK–AlgX alginate export protein complex (expressed in E. coli) | Pseudomonas putida | X-ray diffraction | 2.46 Å | 7ULA |
| P pilus usher translocation domain, PapC130-640 (expressed in E. coli) | Escherichia coli | X-ray diffraction | 3.20 Å | 2VQI |
| P pilus usher translocation domain, PapC146-637 (expressed in E. coli) | Escherichia coli | X-ray diffraction | 3.15 Å | 3FIP |
| P pilus FimD usher bound to FimC:FimH substrate (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.80 Å | 3RFZ |
| P pilus FimD usher in complex with FimC:FimF:FimG:FimH (expressed in E. coli) | Escherichia coli | X-ray diffraction | 3.80 Å | 4J3O |
| P pilus assembly intermediate (FimD-FimC-FimF-FimG-FimH), conformer 1 (expressed in E. coli) | Escherichia coli | Cryo-EM single particle analysis | 4.00 Å | 6E14 |
| P pilus tip assembly intermediate PapC-PapD-PapK-PapF-PapG (expressed in E. coli) | Escherichia coli | Cryo-EM single particle analysis | 7.60 Å | 7LHI |
| Transferrin binding protein A (TbpA) in complex with human transferrin (expressed in E. coli) | Neisseria meningitidis | X-ray diffraction | 2.60 Å | 3V8X |
| Wzi outer-membrane lectin | Escherichia coli | X-ray diffraction | 2.64 Å | 2YNK |
| Opa60 for receptor-mediated engulfment, EXPLOR refined (expressed in Neisseria gonorrhoeae) | Neisseria gonorrhoeae | Solution NMR | / | 2MLH |
| CsgG bacterial amyloid secretion channel (expressed in E. coli) | Escherichia coli | X-ray diffraction | 3.59 Å | 4UV3 |
| CsgG bacterial amyloid secretion channel (expressed in E. coli) | Escherichia coli | X-ray diffraction | 3.10 Å | 4Q79 |
| CsgG-CsgF complex involved in curli biogenesis (expressed in E. coli) | Escherichia coli | Cryo-EM single particle analysis | 2.94 Å | 6LQH |
| CsgG-CsgF complex involved in curli biogenesis (expressed in E. coli) | Escherichia coli | Cryo-EM single particle analysis | 3.38 Å | 6L7A |
| CsgG-CsgF complex involved in curli biogenesis (expressed in E. coli) | Escherichia coli | Cryo-EM single particle analysis | 3.40 Å | 6SI7 |
| FusA plant-ferredoxin receptor (expressed in E. coli) | Pectobacterium atrosepticum | Cryo-EM single particle analysis | 3.20 Å | 4ZGV |
| Omp33 outer membrane protein (expressed in E. coli) | Acinetobacter baumannii | X-ray diffraction | 2.10 Å | 6GIE |
| FrpB TonB-dependent iron transporter (expressed in E. coli) | Neisseria meningitidis | X-ray diffraction | 2.40 Å | 4AIP |
| FhuE TonB-dependent transporter (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.00 Å | 6E4V |
| YncD TonB-dependent transporter (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.96 Å | 6V81 |
| PfeA ferric enterobactin receptor, apo form (expressed in E. coli) | Pectobacterium atrosepticum | X-ray diffraction | 2.12 Å | 5M9B |
| PfeA ferric enterobactin receptor in complex with TCV (expressed in E. coli) | Pseudomonas aeruginosa | X-ray diffraction | 2.57 Å | 5NC3 |
| BcsC cellulose synthase outer membrane channel (expressed in E. coli) | Escherichia coli | X-ray diffraction | 1.85 Å | 6TZK |
| ZnuD zinc transporter (expressed in E. coli) | Neisseria meningitidis | X-ray diffraction | 3.20 Å | 4RDT |
| MtrE Outer Membrane Channel (expressed in E. coli) | Neisseria gonorrhoeae | X-ray diffraction | 3.29 Å | 4MT0 |
| LetB lipophilic Envelope-spanning Tunnel B, model 1 (expressed in E. coli) | Escherichia coli | Cryo-EM single particle analysis | 3.46 Å | 6V0C |
| PcoB copper transporter (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.00 Å | 7PGE |
Table 1. Structural Research of Beta-Barrel Membrane Proteins: Monomeric/Dimeric.
Creative Biostructure is a reliable provider of structural biology services that enable the determination of the high-resolution structures of beta-barrel membrane proteins. Our team of expert scientists utilizes state-of-the-art techniques such as X-ray crystallography, cryo-electron microscopy, and nuclear magnetic resonance spectroscopy to provide clients with accurate structural data. We offer a comprehensive range of services that include protein expression and purification, crystallization, structure determination, and analysis. In addition, our team of experienced scientists can perform high-quality NMR experiments to provide information on protein dynamics and interactions with ligands or other proteins.
If you are interested in leveraging the power of structural biology to explore the monomeric and dimeric structure of beta-barrel membrane proteins, please contact us today. Our custom-designed solutions are tailored to meet the unique requirements of each client and are backed by our commitment to quality and customer satisfaction.
References
- Budiardjo S J, et al. Colicin E1 opens its hinge to plug TolC. Elife. 2022, 11: e73297.
- Schubeis T, et al. A β-barrel for oil transport through lipid membranes: Dynamic NMR structures of AlkL. Proceedings of the National Academy of Sciences. 2020, 117(35): 21014-21021.
- Isom G L, et al. LetB structure reveals a tunnel for lipid transport across the bacterial envelope. Cell. 2020, 181(3): 653-664. e19.