Membrane Protein-Protein Interactions
Membrane proteins play critical roles in various cellular processes, including signal transduction, transport, and cell-cell communication. One of the key aspects of membrane protein function is their ability to interact with other proteins. These interactions are crucial for the assembly and regulation of protein complexes that execute a wide range of cellular functions.
What are Membrane Protein-Protein Interactions?
Membrane protein-protein interactions refer to the specific and transient or permanent associations between membrane-spanning proteins and other proteins, which may be membrane-bound or cytosolic. These interactions are fundamental to a multitude of cellular processes. For instance, G protein-coupled receptors (GPCRs) interact with G proteins to transmit extracellular signals into intracellular responses, while receptor tyrosine kinases (RTKs) dimerize and autophosphorylate to initiate signaling cascades.
1. Direct Interactions: Membrane proteins can interact directly through specific binding sites. These sites are often found in the extracellular, transmembrane, or cytoplasmic domains of the proteins. For example, the interaction between integrins and extracellular matrix (ECM) proteins involves the direct binding of ECM motifs to integrin receptors.
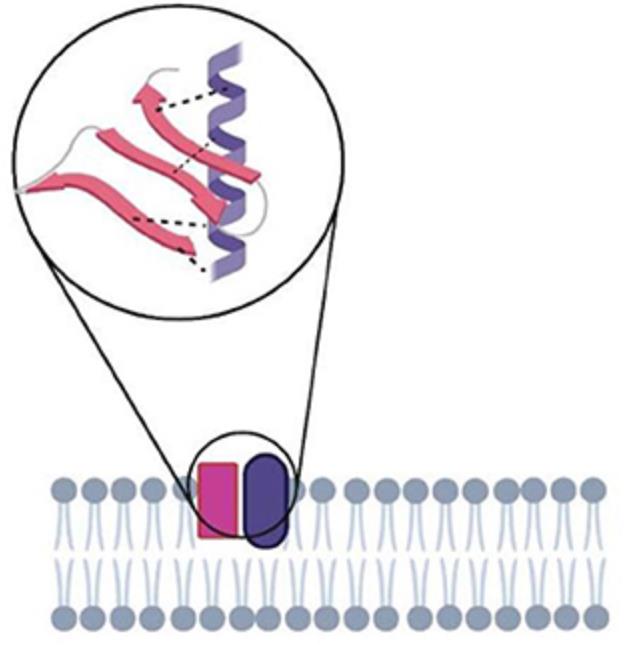 Figure 1. Dynamic membrane protein interactions. (Bagheri Y, et al., 2020)
Figure 1. Dynamic membrane protein interactions. (Bagheri Y, et al., 2020)
2. Indirect Interactions: Indirect interactions involve intermediary proteins or cofactors. For example, scaffold proteins mediate interactions between receptors and downstream signaling molecules, organizing them into efficient signaling hubs. During T-cell signaling, the binding of the MHCp ligand triggers dynamic interactions between the T-cell receptor (TCR) and CD4.
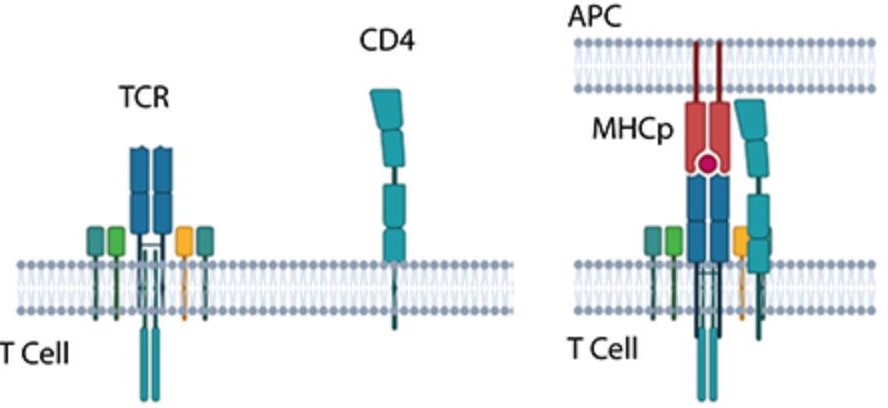 Figure 2. Dynamic TCR-CD4 interactions in T-cell signaling. (Bagheri Y, et al., 2020)
Figure 2. Dynamic TCR-CD4 interactions in T-cell signaling. (Bagheri Y, et al., 2020)
3. Lipid Mediated Interactions: The lipid environment of the membrane can facilitate or inhibit protein interactions. Certain lipids, like cholesterol, can modulate the affinity between membrane proteins by altering the local membrane environment or by directly binding to protein interfaces.
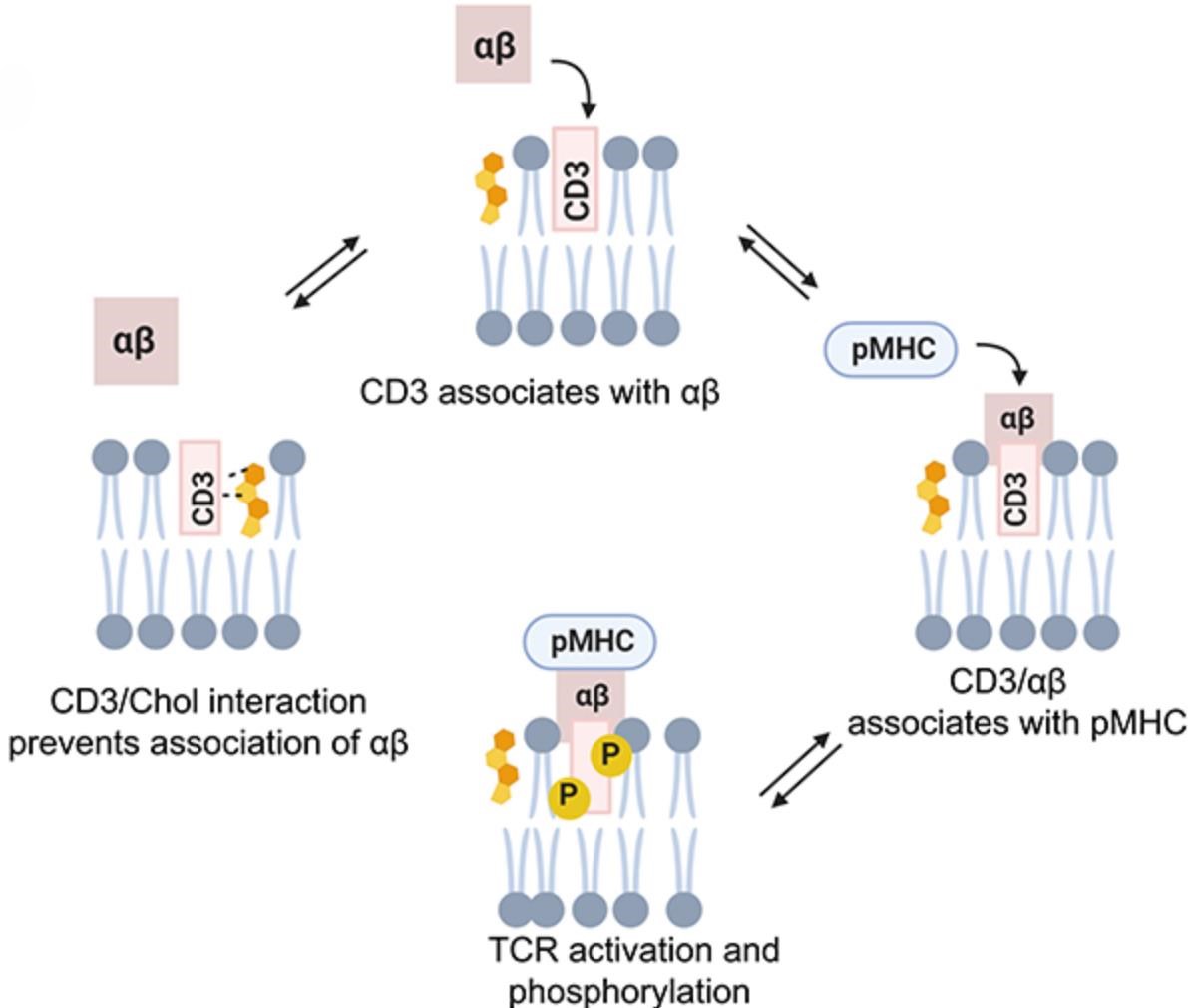 Figure 3. Cholesterol dynamics modulate TCR activation to avoid nonspecific T-cell responses. (Bagheri Y, et al., 2020)
Figure 3. Cholesterol dynamics modulate TCR activation to avoid nonspecific T-cell responses. (Bagheri Y, et al., 2020)
Methods to Study Membrane Protein-Protein Interactions
| Categories | Techniques | Description |
| Biochemical Techniques | Co-Immunoprecipitation (Co-IP) | Co-IP is a widely used technique to study protein-protein interactions. It involves the isolation of a protein complex using a specific antibody against a target protein and identifying the interacting partners through mass spectrometry or immunoblotting. Despite its usefulness, Co-IP has limitations, including the requirement for good antibodies and the potential disruption of weak or transient interactions during the lysis process. |
| Cross-Linking | Cross-linking reagents covalently bond interacting proteins, capturing transient interactions that might be missed by other methods. This approach is combined with mass spectrometry to identify cross-linked peptides, revealing interaction sites and providing spatial constraints that aid in structural modeling. Chemical cross-linkers, like glutaraldehyde, are commonly used, though advancements in photo-cross-linking and click chemistry have improved specificity and reduced off-target effects. | |
| Biophysical Techniques | Fluorescence Resonance Energy Transfer (FRET) | FRET measures the energy transfer between two fluorophores in close proximity (<10 nm), making it ideal for studying protein interactions. It involves labeling interacting proteins with donor and acceptor fluorophores. When the donor fluorophore is excited, energy transfer to the acceptor fluorophore occurs if they are close enough, indicative of interaction. FRET efficiency provides quantitative information about the distance and interaction dynamics. |
| Surface Plasmon Resonance (SPR) | SPR detects changes in refractive index near the sensor surface where a protein is immobilized. When an interacting partner binds, the refractive index changes, providing real-time data on binding kinetics, affinities, and interaction specificity. SPR is highly sensitive and suitable for studying a wide range of interactions, including weak and transient ones. | |
| Nuclear Magnetic Resonance (NMR) Spectroscopy | NMR spectroscopy provides detailed information about protein structure and dynamics in solution. While challenging for large membrane proteins, recent advances in isotopic labeling and sample preparation have enabled the study of smaller membrane proteins and their interactions. NMR can reveal interaction sites and conformational changes upon binding. | |
| Advanced Imaging Techniques | Total Internal Reflection Fluorescence (TIRF) Microscopy | TIRF microscopy allows the visualization of protein interactions at the cell membrane with high spatial and temporal resolution. It excites fluorophores in a narrow region adjacent to the cover glass, minimizing background fluorescence and enabling the study of interactions in the native membrane environment. |
| Super-Resolution Microscopy | Techniques like STORM and PALM surpass the diffraction limit of conventional light microscopy, providing nanometer-scale resolution. These methods are particularly valuable for studying the organization and interactions of membrane proteins in situ. |
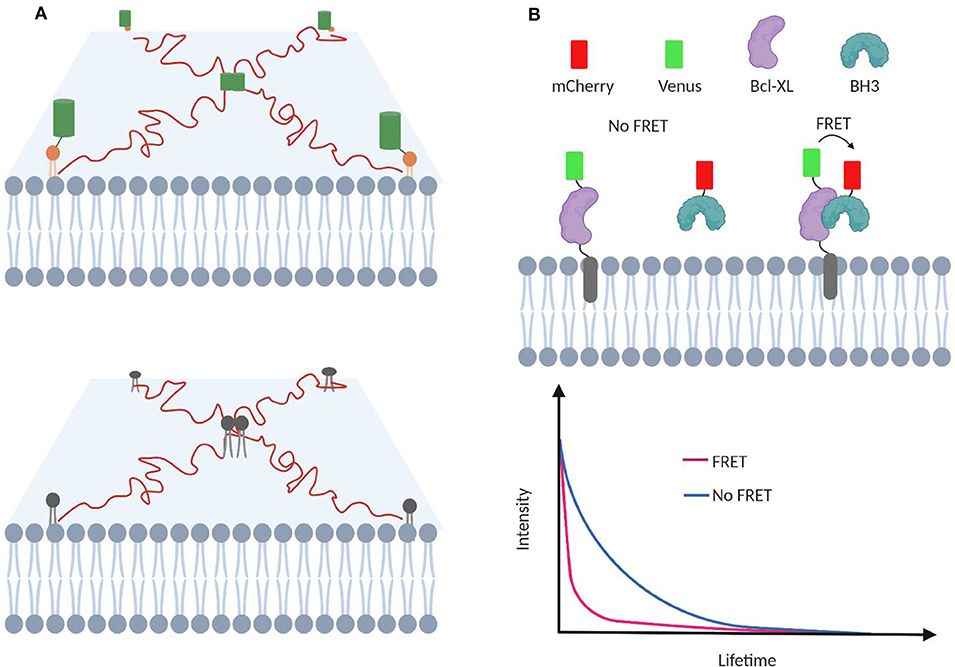 Figure 4. TIRF microscopy for membrane transient interaction analysis. (Bagheri Y, et al., 2020)
Figure 4. TIRF microscopy for membrane transient interaction analysis. (Bagheri Y, et al., 2020)
Current Frontiers and Technological Advances
Structural Insights and High-Resolution Techniques
- Cryo-Electron Microscopy (Cryo-EM): This technique has revolutionized the structural biology of membrane proteins, allowing for the visualization of large protein complexes at near-atomic resolution. Recent advances in cryo-EM have provided detailed structures of GPCRs, ion channels, and other pivotal membrane proteins in complex with their interacting partners.
- Single-Molecule Techniques: Techniques such as single-molecule FRET (smFRET) and atomic force microscopy (AFM) have enabled the study of membrane protein-protein interactions at an unprecedented spatial and temporal resolution. These methods provide insights into the dynamic nature of protein interactions and conformational changes.
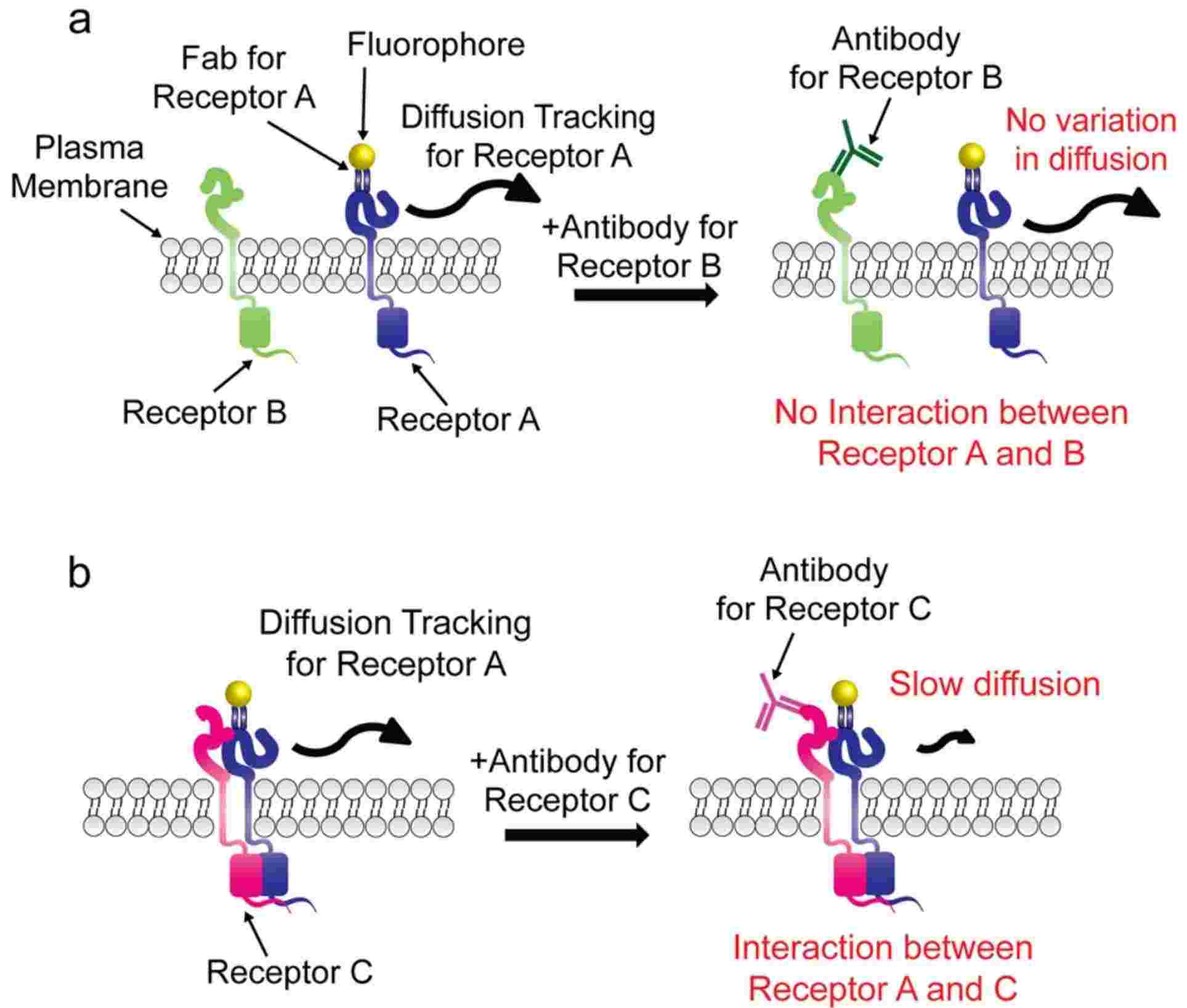 Figure 5. Modified smDIMSA for transient membrane protein interaction analysis. (Jeong M G, et al., 2021)
Figure 5. Modified smDIMSA for transient membrane protein interaction analysis. (Jeong M G, et al., 2021)
Computational Approaches
- Molecular Dynamics (MD) Simulations: MD simulations help understand the dynamic behavior of membrane proteins and their interactions. They provide a detailed understanding of the conformational flexibility and interaction mechanisms at an atomic level.
- Bioinformatics Tools: Various in silico tools and databases, like STRING and BioGRID, predict and analyze protein interaction networks. Machine learning algorithms are increasingly being used to predict membrane protein-protein interactions based on sequence and structural data.
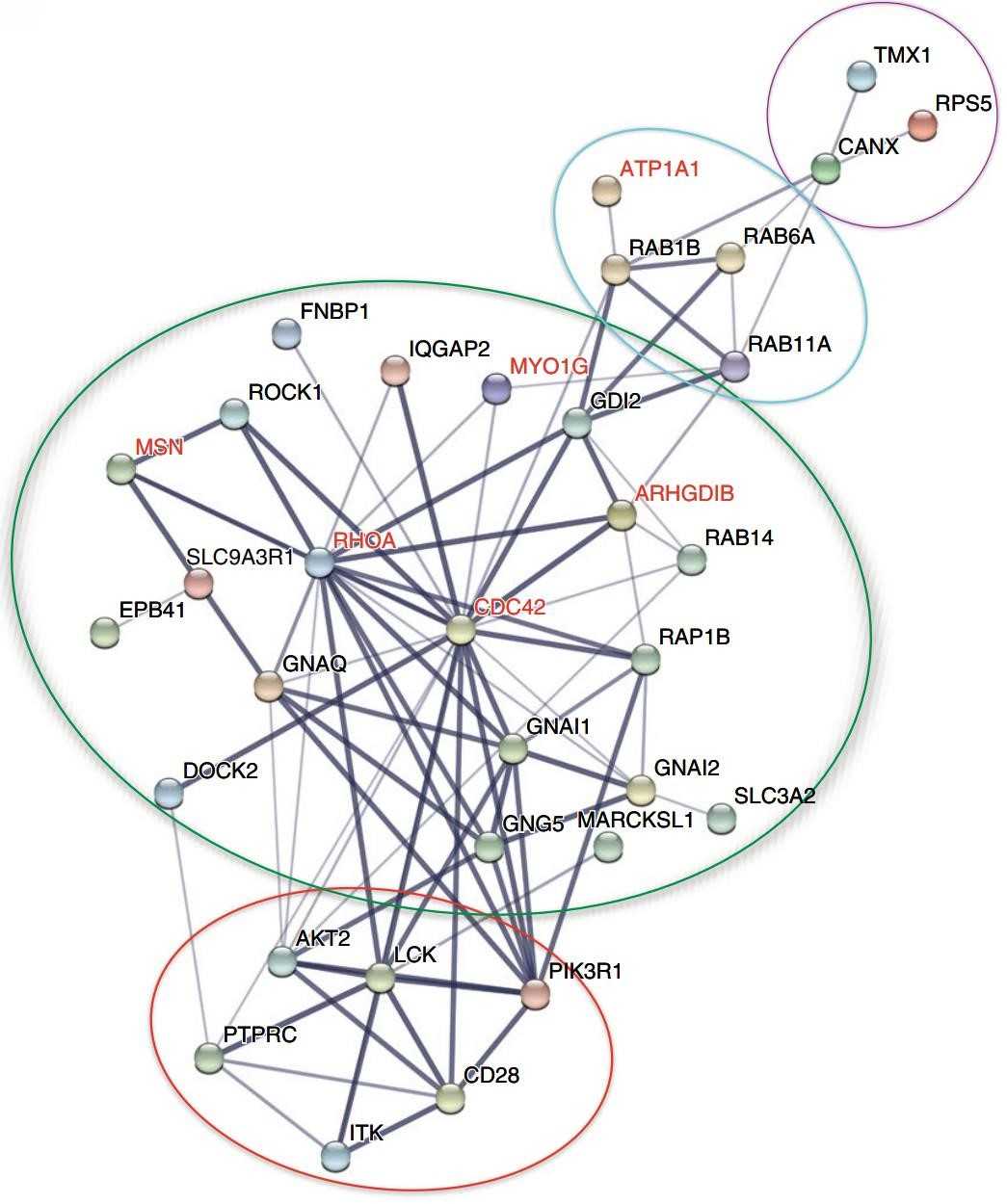 Figure 6. Gene ontology analysis of CD28-interaction proteins using software STRING. (Liu Q, et al., 2018)
Figure 6. Gene ontology analysis of CD28-interaction proteins using software STRING. (Liu Q, et al., 2018)
Functional Studies and In Vivo Techniques
- Optogenetics: This technique enables the precise control of protein interactions using light. Optogenetic tools can activate or inhibit protein interactions in specific cellular contexts, providing a powerful approach to study the physiological roles of membrane protein-protein interactions.
- CRISPR/Cas9: Genome editing through CRISPR/Cas9 allows the creation of specific mutations or tags in membrane proteins, facilitating the study of their interactions and functions in living cells.
Frontier Challenges and Prospects
Heterogeneity of Membrane Environments
Membrane proteins are embedded in lipid bilayers, which are heterogeneous and dynamic. Understanding how the lipid environment influences membrane protein-protein interactions is a significant challenge. Advanced imaging and lipidomics approaches are being developed to dissect these complex interactions.
Transient and Weak Interactions
Many biologically relevant interactions are transient and weak, making them difficult to detect and study. Techniques like cross-linking mass spectrometry and single-molecule approaches are improving our ability to capture and analyze these fleeting interactions.
Integration of Multi-Omics Data
Integrating proteomics, lipidomics, genomics, and interactomics data provides a more comprehensive understanding of membrane protein-protein interactions. Computational tools and machine learning models are essential for managing and interpreting these large datasets.
Therapeutic Targeting of Membrane Protein-Protein Interactions
Disrupting pathological membrane protein-protein interactions offers therapeutic potential for diseases like cancer, neurodegenerative disorders, and infectious diseases. Developing small molecules or biologics that specifically target membrane protein-protein interactions is an emerging area of drug discovery.
At Creative Biostructure, we offer a diverse range of advanced technology services to study membrane protein-protein interactions. Our expertise such as cryo-EM, TIRF microscopy, super-resolution microscopy, FRET and computational modeling, facilitating in-depth analysis and insights into protein dynamics and interactions. Partner with us to leverage cutting-edge methodologies for your research and accelerate your scientific discoveries. If you are interested in our solutions, please feel free to contact us for more information.
References
- Liu Q, Zheng J, Sun W, et al. A proximity-tagging system to identify membrane protein–protein interactions. Nature Methods. 2018. 15(9): 715-722.
- Bagheri Y, Ali A A, You M. Current methods for detecting cell membrane transient interactions. Frontiers in Chemistry. 2020. 8: 603259.
- Jeong M G, Zhou K, Park S, et al. Analysis of transient membrane protein interactions by single-molecule diffusional mobility shift assay. Experimental & Molecular Medicine. 2021, 53(2): 291-299.
- Jiang Y, Thienpont B, Sapuru V, et al. Membrane-mediated protein interactions drive membrane protein organization. Nature Communications. 2022, 13(1): 7373.