Interactions between Membrane Lipids and Membrane Proteins
Membrane lipids and membrane proteins represent two fundamental components of cellular membranes, acting in unison to maintain the dynamic structure and multifaceted functionality essential for cellular life. The cell membrane, composed of a lipid bilayer interspersed with membrane proteins, is a dynamic and complex structure that serves as a barrier and a platform for numerous cellular activities. Membrane lipids, primarily phospholipids and cholesterol, provide the fluidity and flexibility necessary for membrane function, while membrane proteins carry out a variety of roles, including transport, signaling, and cell recognition.
The Structural Foundation: Lipid Bilayer and Membrane Proteins
Lipid Bilayer Composition
The lipid bilayer is primarily composed of amphipathic lipids, such as phospholipids, glycolipids, and cholesterol. These molecules spontaneously arrange themselves into a bilayer with hydrophobic tails tucked inward, away from water, and hydrophilic heads facing the aqueous environments on either side. This arrangement forms a semi-permeable barrier that is fluid yet structurally coherent.
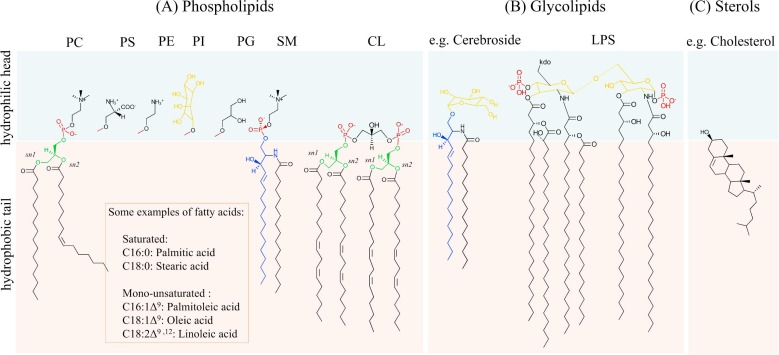 Figure 1. Diversity of lipids in cellular membranes. (Pichler H, et al., 2018)
Figure 1. Diversity of lipids in cellular membranes. (Pichler H, et al., 2018)
Types of Membrane Proteins
Membrane proteins are classified broadly into integral (transmembrane) proteins and peripheral proteins. Integral proteins span the lipid bilayer, typically via alpha-helical or beta-barrel regions, anchoring them firmly within the membrane structure. In contrast, peripheral proteins are associated with the lipid bilayer's surface, adhering through non-covalent interactions with integral proteins or lipid molecules.
Impact of Lipid Bilayer on Protein Function and Structure
Membrane Protein Folding and Stability
The lipid bilayer plays a critical role in the folding and stability of membrane proteins. Hydrophobic interactions between lipid tails and protein domains stabilize the protein structure within the membrane. Proteins such as receptors, channels, and enzymes rely on proper integration within the bilayer to attain their functional conformation.
Transmembrane domains often exhibit a preference for certain lipid environments. For example, the presence of specific phospholipids can enhance or inhibit the proper folding of integral membrane proteins. Misfolded proteins or unstable structures can result in dysfunctional signaling pathways, demonstrating the importance of the lipid environment.
Conformational Dynamics
The fluid nature of the lipid bilayer allows membrane proteins to undergo conformational changes essential for their function. For instance, ion channels and transporters change shape to facilitate the movement of molecules across the membrane. Lipids surrounding these proteins can modulate these conformational shifts, either stabilizing certain states or making transitions more energetically favorable.
Lipid-Protein Specific Interactions
Certain lipids interact specifically with membrane proteins through defined binding sites. These specific interactions can significantly influence protein function. An example is the interaction of phosphoinositides with membrane proteins involved in cell signaling and membrane trafficking. The binding of specific lipids can activate or inhibit protein function, often acting as a regulatory mechanism.
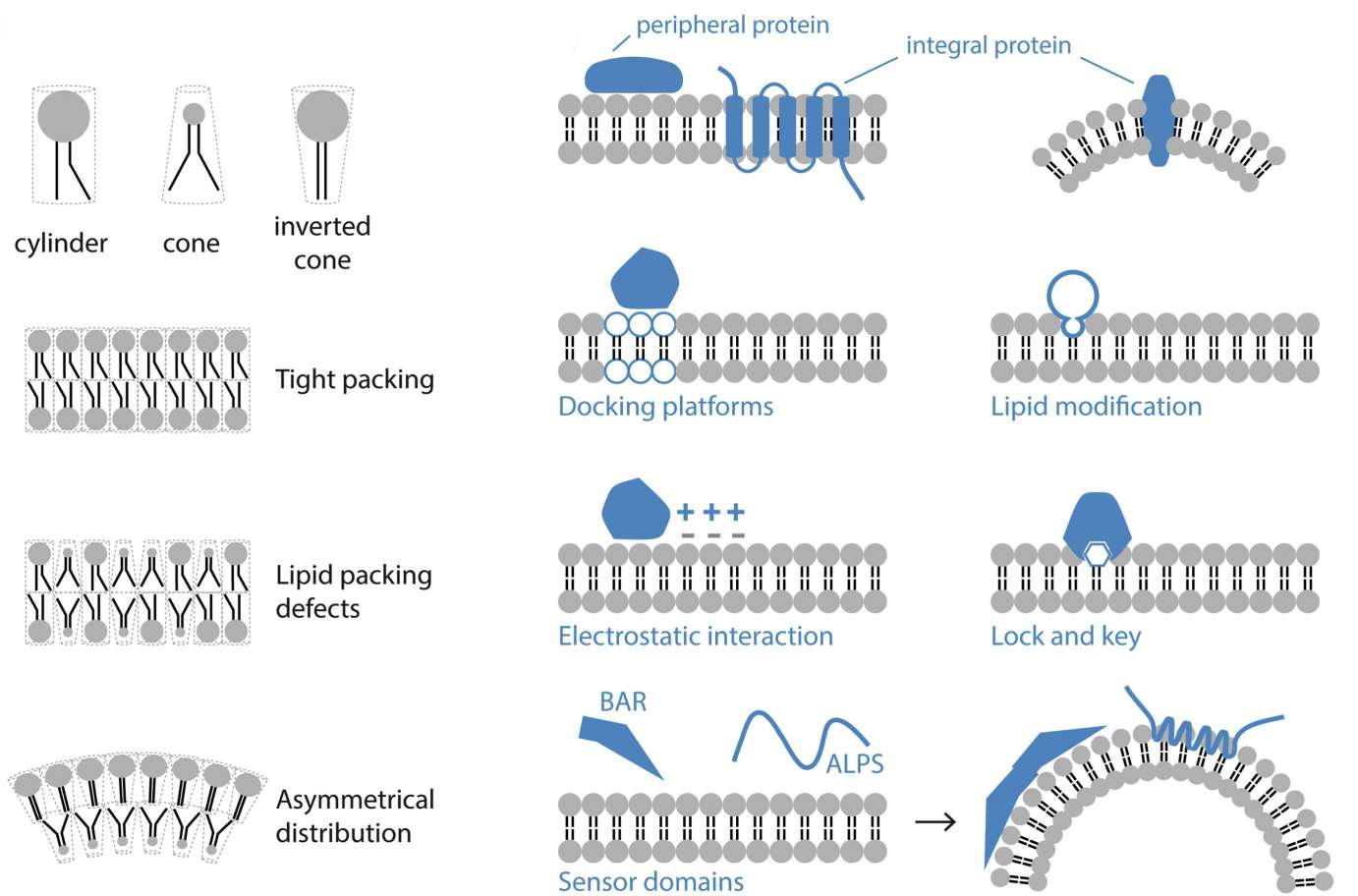 Figure 2. Phospholipids arrange into bilayers with tails inward and heads in water, affecting membrane shape based on their shape. Proteins bind to membranes via lipid attachments, charges, or specific lipid recognition. (de la Ballina L R, et al., 2020)
Figure 2. Phospholipids arrange into bilayers with tails inward and heads in water, affecting membrane shape based on their shape. Proteins bind to membranes via lipid attachments, charges, or specific lipid recognition. (de la Ballina L R, et al., 2020)
Lipid Microdomains: Platforms for Protein Aggregation and Signaling
Definition and Characteristics
Lipid microdomains, often referred to as lipid rafts, are small, dynamic, and heterogeneous regions within the lipid bilayer. They are enriched with cholesterol, sphingolipids, and certain proteins, creating an environment distinct from the surrounding membrane. These microdomains are more ordered and tightly packed, contributing to their unique physical properties.
Functional Implications
Lipid rafts are platforms for the clustering of specific proteins, facilitating interactions that are crucial for cellular signaling. Proteins within these microdomains often have roles in signal transduction, membrane trafficking, and cytoskeletal organization. The assembly of signaling molecules within lipid rafts ensures the spatial and temporal fidelity of signal transmission.
Protein Aggregation
Lipid rafts promote the aggregation of specific membrane proteins, which can be vital for their function. For instance, immune receptors aggregate within lipid rafts upon antigen binding, initiating downstream signaling pathways essential for immune responses. The segregation of proteins into these microdomains can also prevent unintended interactions, maintaining signal specificity.
Signal Transduction
Certain signaling pathways are reliant on the formation and maintenance of lipid rafts. For example, the activation of the T-cell receptor (TCR) involves its localization to lipid rafts, allowing the recruitment of downstream signaling molecules such as Lck, LAT, and SLP-76. Disruption of raft integrity can impair signal transduction, highlighting the importance of these microdomains in cellular communication.
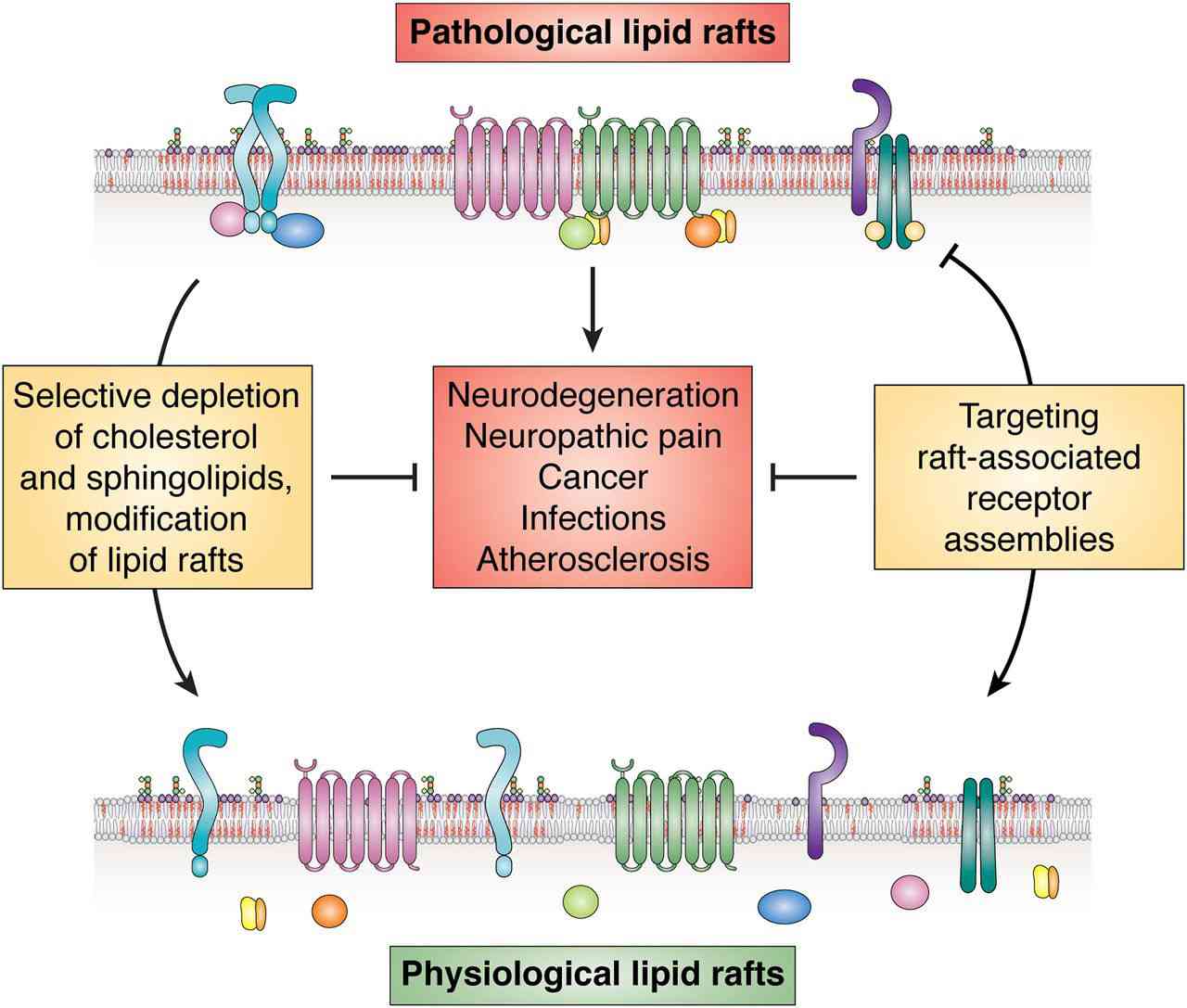 Figure 3. Lipid rafts as a therapeutic target. (Sviridov D, et al., 2020)
Figure 3. Lipid rafts as a therapeutic target. (Sviridov D, et al., 2020)
Interplay between Lipids and Proteins in Membrane Dynamics
Membrane Fluidity and Protein Mobility
Membrane fluidity, governed by lipid composition, affects protein mobility and interactions. A fluid membrane allows proteins to diffuse laterally, facilitating interactions and functional partnerships. Cholesterol and other lipids modulate membrane fluidity, impacting how membrane proteins associate and disassociate during cellular processes.
Lipid-Dependent Protein Sorting
Certain lipids act as molecular markers for protein sorting within cellular membranes. Glycosylphosphatidylinositol (GPI)-anchored proteins, for example, are typically sorted into lipid rafts. This lipid-dependent sorting mechanism is essential for the proper localization and function of proteins within the cell, influencing processes such as endocytosis and exocytosis.
Lipid-Driven Membrane Remodeling
The lipid composition of membranes can drive morphological changes necessary for cellular functions, such as membrane fusion and fission events. Proteins involved in these processes, such as SNAREs, dynamins, and BAR domain-containing proteins, interact with specific lipids to facilitate membrane remodeling. The local lipid environment thus dictates the efficiency and directionality of these critical cellular events.
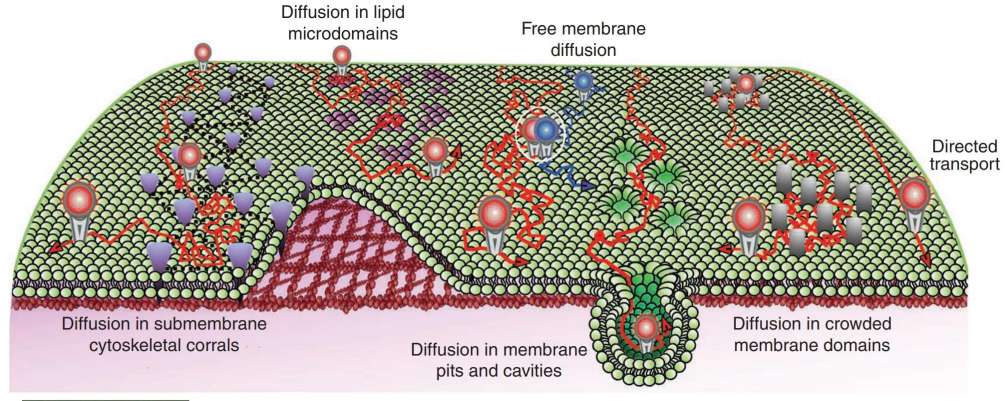 Figure 4. Schematic representation of diverse lateral diffusion for plasma membrane lipids and proteins. Constrained membrane diffusion can be induced by a variety of structures and microdomains, including submembranous skeleton corrals, lipid microdomains, membrane pits and cavities, crowded membrane domains or sites of protein/protein interactions (white circle). (Pinaud F, et al., 2010)
Figure 4. Schematic representation of diverse lateral diffusion for plasma membrane lipids and proteins. Constrained membrane diffusion can be induced by a variety of structures and microdomains, including submembranous skeleton corrals, lipid microdomains, membrane pits and cavities, crowded membrane domains or sites of protein/protein interactions (white circle). (Pinaud F, et al., 2010)
Adcanved Technologies for Studying Membrane Lipid-Protein Interactions
| Techniques | Description | Application |
| Atomic Force Microscopy (AFM) | AFM is a type of scanning probe microscopy that provides high-resolution imaging of biological samples under physiological conditions. | It can visualize the topography of membrane proteins and lipids, and measure the forces between membrane components to understand their interactions. |
| Cryo-Electron Microscopy (Cryo-EM) | Cryo-EM involves freezing biological samples in vitreous ice and visualizing them using electron microscopy. | It offers high-resolution structures of membrane proteins in complex with lipids, illuminating how lipids are arranged around proteins. |
| Nuclear Magnetic Resonance (NMR) Spectroscopy | NMR spectroscopy uses the magnetic properties of nuclei to determine the physical and chemical properties of atoms or molecules. | Solid-state NMR can be used to study the dynamics and interactions of membrane proteins with lipids in their native environments. |
| Mass Spectrometry (MS) | MS measures the mass-to-charge ratio of ions to identify and quantify molecules. | MS can be used to analyze the lipidomics of membrane samples and the lipid binding sites on membrane proteins. |
| Fluorescence Resonance Energy Transfer (FRET) | FRET is a distance-dependent interaction between two light-sensitive molecules (a donor and an acceptor). | It can investigate the proximity and interaction between lipids and membrane proteins by labeling them with suitable fluorophores. |
| Molecular Dynamics (MD) Simulations | MD simulations use computational methods to model the movement of atoms and molecules over time. | These simulations can predict the behavior of membrane lipids and proteins, and provide insights into their interaction mechanisms at atomic resolution. |
| Lipid Nanodiscs | Nanodiscs are small, disc-shaped membrane mimetics that provide a native-like environment for membrane proteins. | They are used to study the functional and structural properties of membrane proteins, along with their lipid interactions. |
| Single-Molecule Techniques | These techniques focus on observing individual molecules rather than ensemble averages. | Techniques like single-molecule FRET or optical tweezers can provide detailed insights into the interactions between single lipid molecules and membrane proteins. |
| Surface Plasmon Resonance (SPR) | SPR measures the binding interactions between molecules on a metal surface by detecting changes in light reflectivity. | It can be used to quantify the binding affinity and kinetics of membrane proteins and lipids. |
| CARS (Coherent Anti-Stokes Raman Scattering) Microscopy | A label-free imaging technique that generates contrast based on the vibrational properties of molecules. | It can be used to image lipids and proteins in their native membrane environment, allowing for the study of their interactions. |
At Creative Biostructure, we offer a wide range of advanced technologies and services for studying the interactions between membrane lipids and membrane proteins. Our state-of-the-art tools such as atomic force microscopy (AFM), cryo-electron microscopy, solid-state NMR, and various biophysical and biochemical assays. By leveraging these cutting-edge methodologies, we can provide comprehensive insights into the structural and functional dynamics of membrane components. Whether for drug discovery, functional analysis, or structural elucidation, our expert team is dedicated to facilitating your research with high precision and reliability. Contact us to learn more about how we can support your study of membrane lipid and protein interactions.
References
- Pinaud F, Clarke S, Sittner A, et al. Probing cellular events, one quantum dot at a time. Nature Methods. 2010. 7(4): 275-285.
- Pichler H, Emmerstorfer-Augustin A. Modification of membrane lipid compositions in single-celled organisms–From basics to applications. Methods. 2018. 147: 50-65.
- de la Ballina L R, Munson M J, Simonsen A. Lipids and lipid-binding proteins in selective autophagy. Journal of Molecular Biology. 2020. 432(1): 135-159.
- Sviridov D, Mukhamedova N, Miller Y I. Lipid rafts as a therapeutic target: thematic review series: biology of lipid rafts. Journal of Lipid Research. 2020. 61(5): 687-695.
- Westra M, Gutierrez Y, MacGillavry H D. Contribution of membrane lipids to postsynaptic protein organization. Frontiers in Synaptic Neuroscience. 2021. 13: 790773.