Screening of Spatially Separated Variants
Creative Biostructure has developed a comprehensive protein evolution and protein engineering platform through years of experience, our seasoned scientists can provide customized screening of spatially separated variants for protein engineering.
Spatial separation (also called clonies screening) of individual mutants is able to preserve the linkage between phenotype and genotype. It is well known that gene variants are expressed in a unicellular model organism such as E. coli that can be screened as colonies on solid media or other culture media (such as liquid culture plate). Spatial separation has a practical throughput limit of fewer than approximately 104 library members per screening round, however, the key advantage of this method is its broad compatibility with many different assay techniques. Spatial separation is the most widely used screening strategy for directed evolution, almost any enzymatic activity or antibody development can be screened in a spatially separated library. In addition, spatial separation is the time-consuming technique, and which has to take the limitations of screened library size into consideration. Creative Biostructure can perform nuclear magnetic resonance (NMR), fluorescent assay, high-performance liquid chromatography (HPLC), gas chromatography or mass spectroscopy (MS) to directly monitor substrate consumption or product formation.
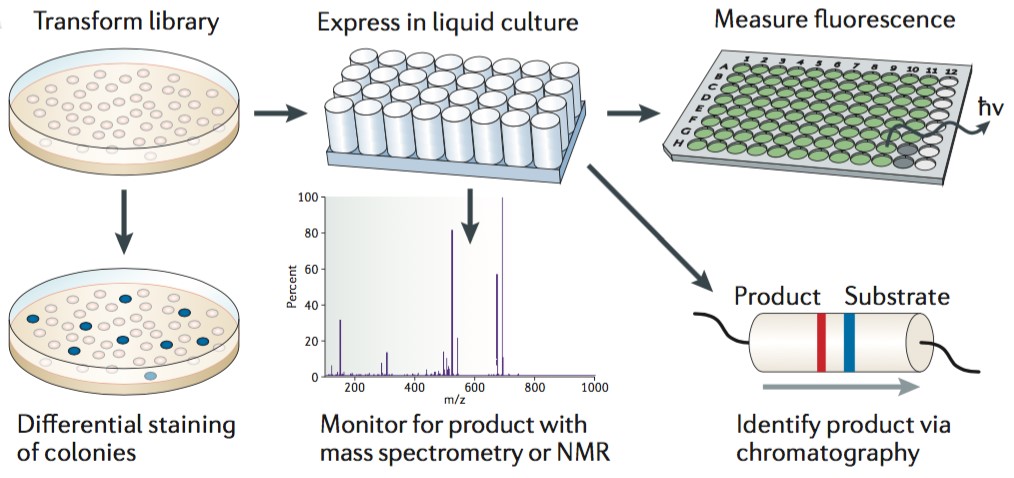
Figure 1. Clonally isolated variants are screened as colonies on solid media or in liquid culture. (Nature Reviews Genetics, 2015)
Screening of spatially separated variants is a low-throughput screen method, which is an understanding of structure-activity relationships within the protein of interest can be necessary to maximize the probability of accessing a desired variant. High-throughput screen relies on the rapid assessment of optical features such as colour, fluorescence, luminescence or turbidity. Most biomolecules are not associated with directly observable phenotypes and thus require a fluorescent, colorimetric or other readily detectable reporter. For instance, fluorescent proteins can offer a readily sreenable phenotype, and this approach is used to Arch rhodopsin, a form of channel rhodopsin to exhibit voltage-dependent fluorescence and applied to directly image neuronal activity. In sum, automated fluorescence technique and robotic colony picking lighten the tedious workload of these screens, but the physical and material constraints associated with spatial separation inherently limit throughput.
Creative Biostructure’s custom screening or selection services can provide you a wide range of first-class strategies for directed evolution. Creative Biostructure also provides Membrane Protein Platform and Phage Display Platform for your specific projects. Please feel free to contact us for a detailed quote.
References:
Blue white screen. (https://en.wikipedia.org/wiki/Blue_white_screen)
M. S. Packer and D. R. Liu (2015). Methods for the directed evolution of proteins. Nature Reviews Genetics, 16: 379-394.