Structural Research of Phospholipid Synthases
Phospholipids are essential for cellular functions such as cell metabolism, signaling, and the immune system. The synthesis of phospholipids involves many enzymes and pathways. X-ray crystallography is a powerful tool for determining the three-dimensional structure of proteins and other biological macromolecules. In recent years, great progress has been made in the structural determination of enzymes involved in phospholipid synthesis, which provides valuable information for understanding the catalytic mechanism and substrate specificity of these enzymes.
Structural approach to bacterial cyclopropane-fatty acyl-phospholipid synthetase (CFAS)
Cyclopropane-fatty acyl-phospholipid synthetase (CFAS) synthesizes cyclopropane fatty acids (CFAs) upon transfer of a methylene group from S-adenosylmethionine (SAM) via the cis-double bond of the unsaturated fatty acyl chain. In the structural study of CFAS, crystallization was first carried out using the hanging drop vapor diffusion method. Diffraction data were collected at the beamline of SSRF. Data processing was performed using the HKL2000 program. The structure was solved by the molecular replacement program PHASER of the CCP4 package in conjunction with the search model. Model construction using ARP/wARP and refinement by REFMAC to obtain the structure.
Structural analysis of CFAS
CFAS protein consists of two structural domains, the N-terminal structural domain, from Glu3 to Lys96, and the C-terminal structural domain, from Ser114 to Leu390. The N-terminal structural domain of CFAS contains four strands of β-sheets, and there is a layer of four α-helices overlying the β-sheets. Between the layer of α-helices and the β-sheets are lipid binding sites. The core region of the C-terminal structural domain of CFAS consists of seven strands of β-sheets flanked by α-helices. Phospholipids are located in λ-shaped tunnels that extend from the surface between the N- and C-terminal structural domains to a large pocket in the center of CFAS.
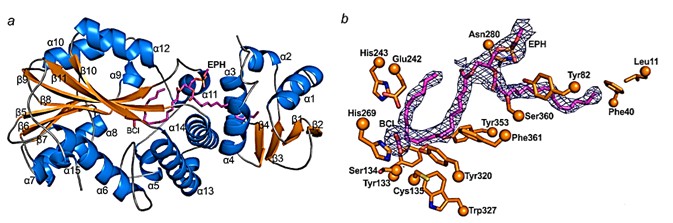 Figure 1. Structure of CFAS. (Ma Y, et al., 2019)
Figure 1. Structure of CFAS. (Ma Y, et al., 2019)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Cyclopropane-fatty-acyl-phospholipid synthase | Lactobacillus acidophilus NCFM | X-ray diffraction | 2.7 Å | 5Z9O |
| Cyclopropane fatty acid synthase | Escherichia coli | X-ray diffraction | 2.073 Å | 6BQC |
| Cyclopropane fatty acid synthase with bound ligands | Aquifex aeolicus VF5 | X-ray diffraction | 1.6 Å | 7QOS |
| Phosphatidylinositol phosphate synthase (PgsA1) in apo form | Mycobacterium tuberculosis H37Rv | X-ray diffraction | 2.9 Å | 6H53 |
| Phosphatidyl serine synthase (PSS) in the closed conformation with bound citrate. | Methanocaldococcus jannaschii DSM 2661 | X-ray diffraction | 2.8 Å | 7B1N |
| Phosphatidyl serine synthase (PSS) in the closed conformation with bound citrate. | Methanocaldococcus jannaschii DSM 2661 | X-ray diffraction | 2.2 Å | 7B1K |
| Phosphatidyl serine synthase (PSS) in transition state. | Methanocaldococcus jannaschii DSM 2661 | X-ray diffraction | 2.51 Å | 7POW |
| phosphatidylserine synthase | Haemophilus influenzae | X-ray diffraction | 2.2 Å | 3HSI |
| Phosphatidylethanolamine-binding protein | Plasmodium vivax | X-ray diffraction | 1.3 Å | 2GZQ |
| Phosphatidylethanolamine binding protein | Bos taurus | X-ray diffraction | 1.84 Å | 1A44 |
| A-PGS | Pseudomonas aeruginosa PAO1 | X-ray diffraction | 2.3 Å | 4V35 |
| L-PGS | Bacillus licheniformis | X-ray diffraction | 2.6 Å | 4V36 |
Table 1. Structural research of phospholipid synthases.
Structural analysis of phospholipid synthase is significant for revealing the regulatory mechanisms during phospholipid formation. The results of structural studies can help to understand the mechanism by which phospholipid synthases are involved in synthetic reactions.
Creative Biostructure is a leading biotechnology company focused on protein structural biology. We provide services related to structural studies of phospholipid synthases and various types of proteins, including NMR spectroscopy, cryo-electron microscopy (cryo-EM) and X-ray crystallography. Our professional team has extensive experience and advanced equipment to provide comprehensive support for your research. If you are interested in protein structure analysis and related research fields, please contact us for more details.
References
- Ma Y, et al. Crystal structure of bacterial cyclopropane-fatty-acyl-phospholipid synthase with phospholipid.J Biochem. 2019.166(2):139-147.
- Centola M, et al. Crystal structures of phosphatidyl serine synthase PSS reveal the catalytic mechanism of CDP-DAG alcohol O-phosphatidyl transferases. Nat Commun. 2021.12(1):6982.