Mempro™ Nanodisc Model Development
Creative Biostructure has developed an advanced and magic Mempro™ Nanodisc platform to provide computational studies of various nanodiscs of different sizes and compositions with/without embedded membrane proteins.
Phospholipid nanodiscs are 10 nm sized discoloidal protein-lipid particles which are consisting of around 150 phospholipids arranged in a central bilayer, stabilized by two amphipathic membrane scaffold proteins that wrap around the rim of the bilayer. Nanodiscs can shield the acyl chains of lipids from the outside aqueous environment, providing a soluble membrane mimetic into which membrane proteins can be incorporated.
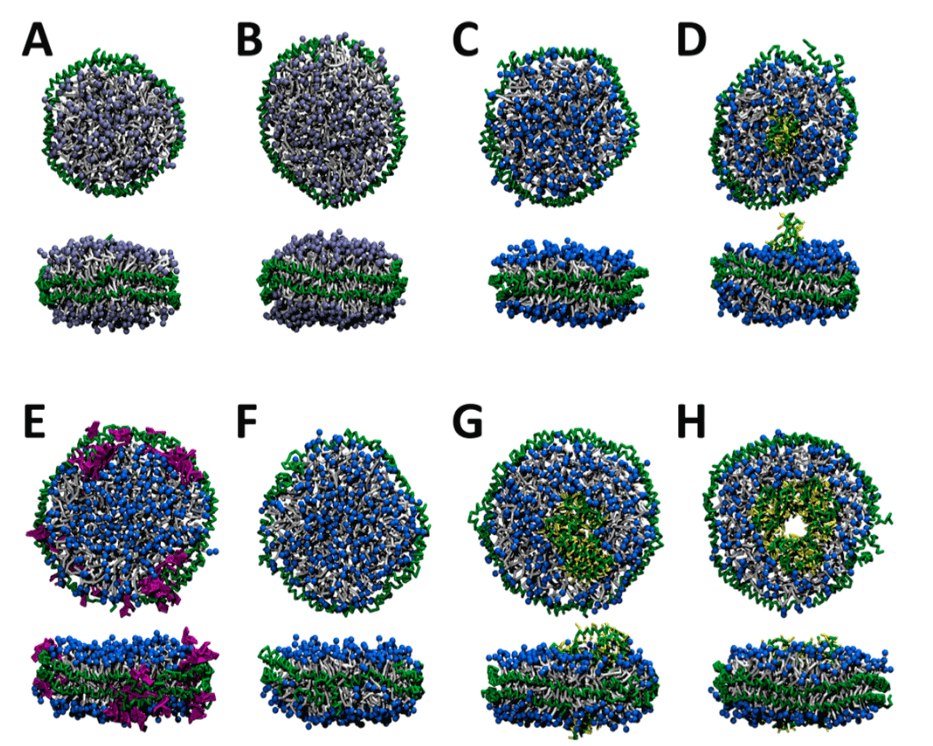 Figure 1. Representative structures from the molecular dynamics simulations of nanodiscs composed of a variety of MSPs and phospholipids. (Journal of Chemical Theory and Computation, 2015)
Figure 1. Representative structures from the molecular dynamics simulations of nanodiscs composed of a variety of MSPs and phospholipids. (Journal of Chemical Theory and Computation, 2015)
Creative Biostructure has applied nanodiscs as a powerful tool to study membrane proteins because they can provide the native-like lipid environment, and we have developed a wide range of research techniques based on biophysics and biochemistry. There are still limited high-resolution structures of nanodiscs with membrane proteins, therefore, molecular dynamics simulation presents a new approach to construct representative models of different nanodiscs. The most significant advantage of computational methods is that the size and composition of nanodisc can be precisely controlled. As the Figure 1 shown, there are different nanodiscs composed of MSP1 and 122 POPC lipids (A), MSP1 and 160 POPC lipids (B), MSP1 and 162 DMPC lipids (C), MSP1, OmpX, and 140 DMPC lipids (D), MSP1E1, 22 GM1 glycolipids, and 204 DMPC lipids (E), MSP1E1 and 206 DMPC lipids (F), MSP1E1, GluT1, and 186 DMPC lipids (G), and MSP1E2, bR trimer, and 130 DMPC lipids (H).
Creative Biostructure is expertized in building novel nanodisc models as membrane mimetics by computational methods. We are able to create MSPs structures of different lengths and build pre-assembled nanodiscs with or without membrane proteins rapidly. Our strategy is to combine both the coarse-grained (CG) simulation and all-atom simulation. There are three major advantages of molecular dynamics simulations:
- 1. Comparison with the experimental data of membrane proteins in nanodiscs directly;
- 2. Interpretation of the experimental data with a detailed structural insight;
- 3. Easy way to test nanodisc-related hypothesis and direct the experiment design.
Creative Biostructure can help you to acquire a variety of properties of nanodiscs by computational studies, including lipid order parameter, the size and shape distribution, area per lipid, and thickness. Molecular dynamics simulations can not only investigate the properties of nanodiscs but also study the structure and function of membrane proteins.
Creative Biostructure also provides a wide range of Mempro™ Nanodisc products and services. Please see the related sections for further information and feel free to contact us for a detailed quote.
References:
A. Debnath, et al. (2015). Structure and Dynamics of Phospholipid Nanodiscs from All-Atom and Coarse-Grained Simulations. Journal of Physical Chemistry B, 119(23): 6991-7002.
M. Vestergaard, et al. (2015). Bicelles and Other Membrane Mimics: Comparison of Structure, Properties, and Dynamics from MD Simulations. Journal of Physical Chemistry B, 119: 15831−15843.
I. Siuda, et al. (2015). Molecular Models of Nanodiscs. Journal of Chemical Theory and Computation, 11: 4923−4932.