Structural Research of Squalene-Hopene Cyclases
Squalene-hopene cyclases (SQHcs) are a class of catalytic enzymes involved in a crucial step in the steroid biosynthesis pathway. They can catalyze the cyclization reaction between glutaric acid and carotenoids, converting linear polycyclic terpenoids into cyclic terpenoids. These enzymes are widely found in organisms such as plants, bacteria, and fungi, and have diverse structures and functions. Structural elucidation of SQHcs is critical for understanding their catalytic mechanisms and biological functions. Understanding the structure of SQHcs can help scientists to reveal its mode of interaction with substrates and further investigate its catalytic mechanism and reaction kinetics. In addition, the role of SQHcs in natural product synthesis can also be studied by structural resolution, which can provide an important reference for biotechnology and drug development.
Protein structure analysis uses a variety of techniques. For example, the structure of SQHcs is resolved by high-resolution X-ray crystallography. Studies such as conformational analysis and kinetic simulations of SQHcs can be performed using computational methods. Using advanced electron microscopy techniques, the overall structure and substrate binding mode of SQHcs can be resolved.
SQHcs is a protein of 70-75 kDa consisting of 631 amino acids and 7 PTFB repeats. A. acidocaldarius SHC structural studies elucidate the biochemical mechanism of the SHC reaction. The crystallized A. acidocaldarius SHC is a homodimer with dimensions of 50 x 70 Å (70 kDa per monomer). Each subunit consists of α-helical structural domains that form a dumbbell-shaped structure. The first domain consists of a regular (α/α) 6-barrel structure, while the second domain shows a less periodic α-barrel structure. A comparison of the two structural domains suggests that the second structural domain may have been formed after gene duplication in the first structural domain. Crystal structure analysis confirmed that SHC is only partially integrated into the membrane and apparently does not span the entire lipid bilayer (unit membrane protein). The membrane-bound portion of the enzyme is a nonpolar region surrounded by positively charged amino acids that force the enzyme to anchor to the negatively charged surface of the phospholipid membrane.
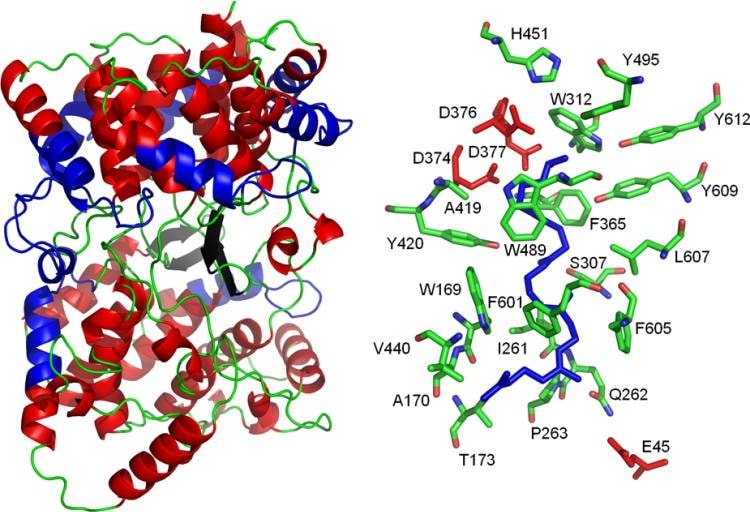 Figure 1. Structure and active site of the A. acidocaldarius SHC. (Siedenburg G, et al., 2011)
Figure 1. Structure and active site of the A. acidocaldarius SHC. (Siedenburg G, et al., 2011)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Squalene-hopene cyclase: Alicyclobacillus acidocaldarius | Alicyclobacillus acidocaldarius | X-ray diffraction | 2.00 Å | 2SQC |
| SQUALENE-HOPENE CYCLASE | Alicyclobacillus acidocaldarius | X-ray diffraction | 2.80 Å | 3SQC |
Table 1. Structural Research of Squalene-Hopene Cyclases.
Creative Biostructure is a biological company focused on protein structural biology research. We offer a variety of protein structural analysis services. Our X-ray crystallography service maximizes protein crystal quality and size by optimizing crystal growth conditions to obtain high-resolution crystallographic structure data. Nuclear magnetic resonance (NMR) services, protein NMR spectroscopy: including secondary structure, tertiary structure, and dynamic properties structural calculation and molecular simulation services for proteins using NMR data, etc. In addition to the above core services, we also provide a variety of services such as protein structure-related customization services, protein structure analysis interpretation and reporting, etc. If you would like to learn more about what we do, feel free to contact us anytime.
Reference
- Siedenburg G, Jendrossek D. Squalene-hopene cyclases. Appl Environ Microbiol. 2011 Jun; 77(12): 3905-15.