What is Exosomal RNA Sequencing?
It has been found that exosomes, as a class of extracellular vesicles containing DNA, RNA, proteins, and lipids, are widely involved in regulating important cellular physiological activities. Exosomal RNAs, especially non-coding RNAs, have obvious expression differences in different disease models and are expected to be important molecular markers for disease diagnosis, personalized treatment, and prognosis, and to reveal the mechanism of disease occurrence. Using high-throughput transcriptome sequencing technology, RNA molecules (e.g. miRNA, lncRNA, circRNA, mRNA) contained in exosomes can be detected for in-depth study of exosomal RNA identification and functional regulation.
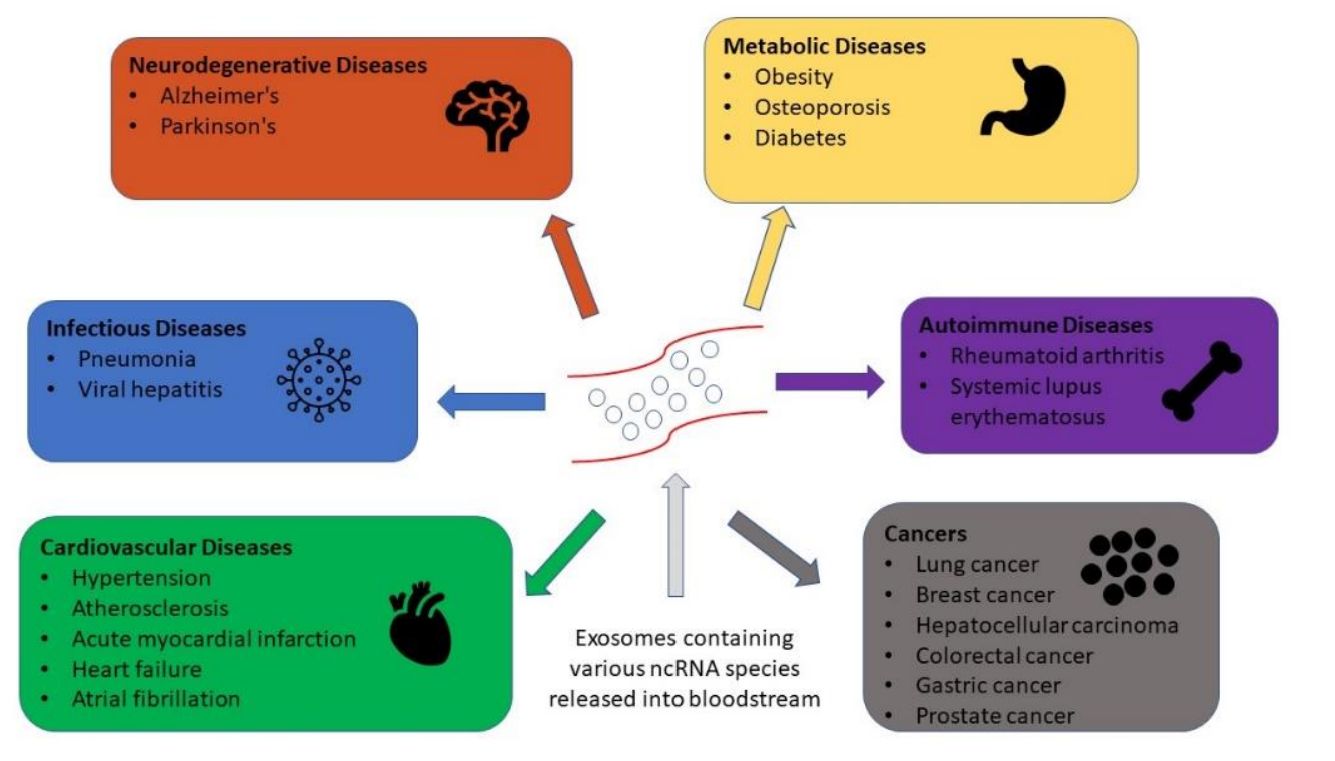 Figure 1. Exosomal RNAs involved in human disease progression. (Elkommos-Zakhary M, et al., 2022)
Figure 1. Exosomal RNAs involved in human disease progression. (Elkommos-Zakhary M, et al., 2022)
Exosome RNA Sequencing Workflow
- Exosome Isolation - The first step in exosome RNA sequencing is to isolate exosomes from biological fluids or cell culture media. This can be accomplished using a variety of techniques such as ultracentrifugation, size exclusion chromatography, or immunoaffinity-based methods.
- RNA Extraction and Purification - After the isolation of the exosomes, the next step is to extract and purify the RNA. we use RNA extraction kits specifically designed to treat extracellular vesicles to ensure the recovery of high-quality RNA suitable for downstream analysis.
- Library Preparation - We convert the RNA molecules into complementary DNA (cDNA) and add junctions or barcodes for sequencing. We choose the appropriate library preparation method based on specific research objectives, such as poly(A) selection, ribosomal RNA depletion, or small RNA enrichment.
- Sequencing - The prepared library is then loaded onto a high-throughput sequencing platform to generate millions of sequencing reads representing exosomal RNA transcripts.
- Bioinformatics Analysis - Raw sequencing data undergoes a series of bioinformatics analyses, including quality control, read matching, transcript quantification, and differential expression analysis, to identify the specific RNA species present in exosomes and their relative abundance.
Exosomal RNA Sequencing Technology Applications
- Biomarker Discovery
In areas such as cancer, neurodegenerative diseases, and cardiovascular diseases, researchers have successfully utilized exosomal RNA sequencing technology to identify biomarkers with diagnostic and predictive value.
- Intercellular Communication Research
Exosomes communicate between cells by delivering signaling molecules. Exosome RNA sequencing technology enables us to study these signaling molecules and gain insight into the mechanisms of intercellular communication.
- Drug Delivery System
Exosomes are often used as drug delivery carriers to increase the therapeutic efficacy of drugs and reduce side effects. Exosome RNA sequencing technology helps to study the behavior and mechanism of action of drugs in exosomes.
- Tumor Microenvironment Research
Exosomes can change the behavior of cells around tumors and promote tumor development. RNA sequencing technology allows a better understanding of the tumor microenvironment and the role of exosomes in it.
Advances in Exosome RNA Sequencing
- Single Exosome RNA Sequencing
Advances in microfluidic and single-cell technologies have driven the development of techniques for sequencing the RNA content of individual exosomes. This approach provides unprecedented resolution and insight into the heterogeneity of exosomal RNA profiles in populations.
- Multi-omics Integration
Integration of exosome RNA sequencing data with other histological techniques, such as proteomics, metabolomics, and genomics, allows for a more comprehensive understanding of the molecular characterization and functional significance of exosomes in a variety of biological and pathological environments.
- Spatial Transcriptomics
The combination of exosomal RNA sequencing and spatial transcriptomics techniques can reveal the local distribution and potential intercellular communication mediated by exosomes in specific tissue microenvironments.
- Artificial Intelligence and Machine Learning
The vast amount of data generated by exosomal RNA sequencing studies has facilitated the application of artificial intelligence and machine learning algorithms to identify complex patterns, predict disease outcomes, and develop personalized treatment strategies.
Cases
- Small RNA Sequencing Analysis of Exosomes in IUGR Lambs
Pregnant ewes in a hot environment may cause intrauterine growth retardation (IUGR) in lambs. Researchers isolate umbilical cord plasma exosomes and screen for differentially expressed miRNAs using small RNA sequencing. Then, predicting the target genes of differentially expressed miRNAs and analyzing the target genes for GO and KEGG pathway enrichment are performed. The results show that oar-miR-411a-5p is significantly down-regulated, while oar-miR-200 is significantly up-regulated. The miRNAs identified in exosomes have the potential to be used as biomarkers for the diagnosis and treatment of IUGR fetuses.
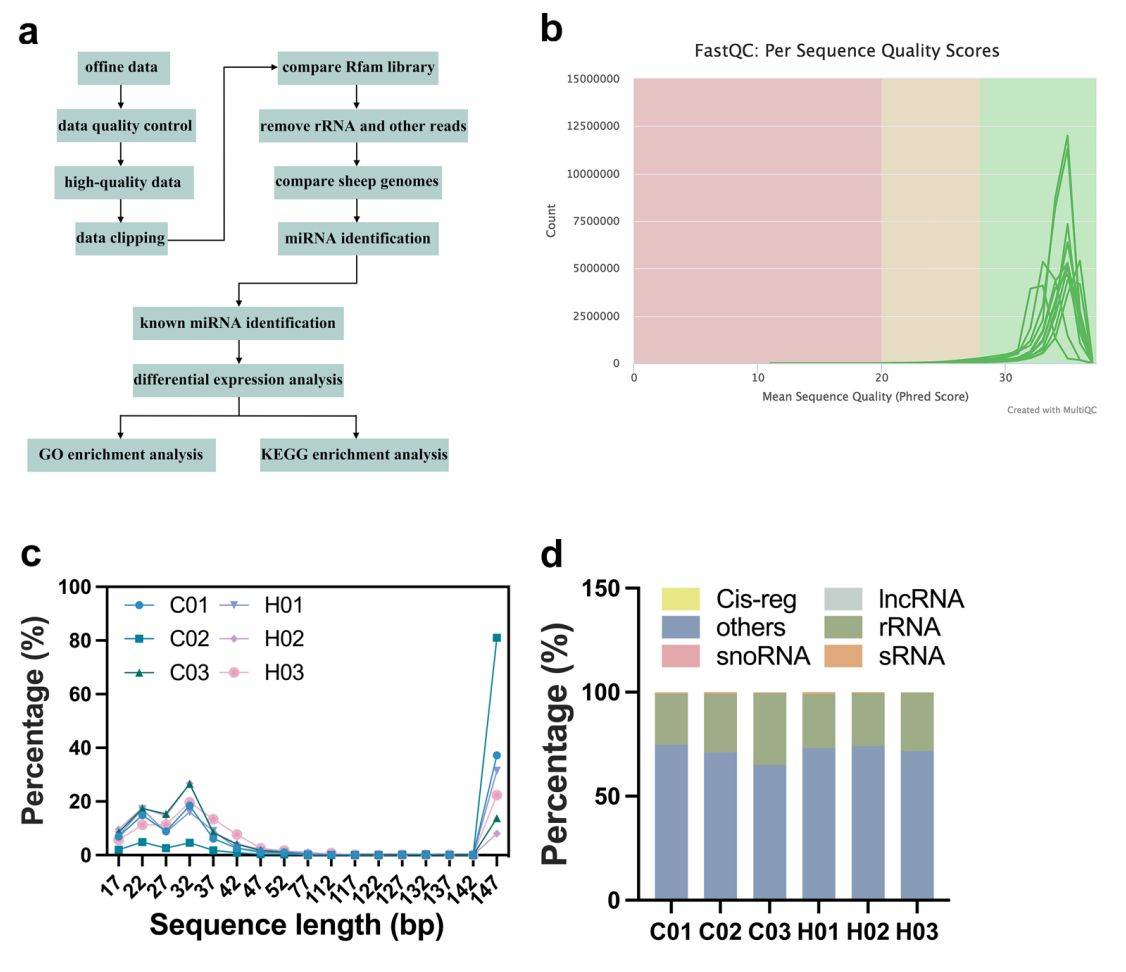 Figure 2. Small RNA sequencing of exosomes from normal and IUGR lambs. (Lu J, et al., 2023)
Figure 2. Small RNA sequencing of exosomes from normal and IUGR lambs. (Lu J, et al., 2023)
- RNA Analysis of Serum Exosomes in Patients with Liver Failure
Hepatitis B is an infectious disease characterized by liver damage. In recent years, researchers have utilized bioinformatics techniques to determine the potential function of exosomal RNA in hepatitis B. Exosomes from healthy control (HC) individuals, patients with chronic hepatitis B (CHB), and patients with hepatitis B virus-induced chronic acute liver failure (HBV-ACLF) have been analyzed by RNA sequencing. Exosomes from CHB and HBV-ACLF patients showed patterns of up-and-down-regulation of differentially expressed genes compared to HC. The findings suggest that RNA cargoes are selectively packaged into exosomes under HBV attack, and these may represent potential targets for diagnosis and treatment of HBV-induced liver injury.
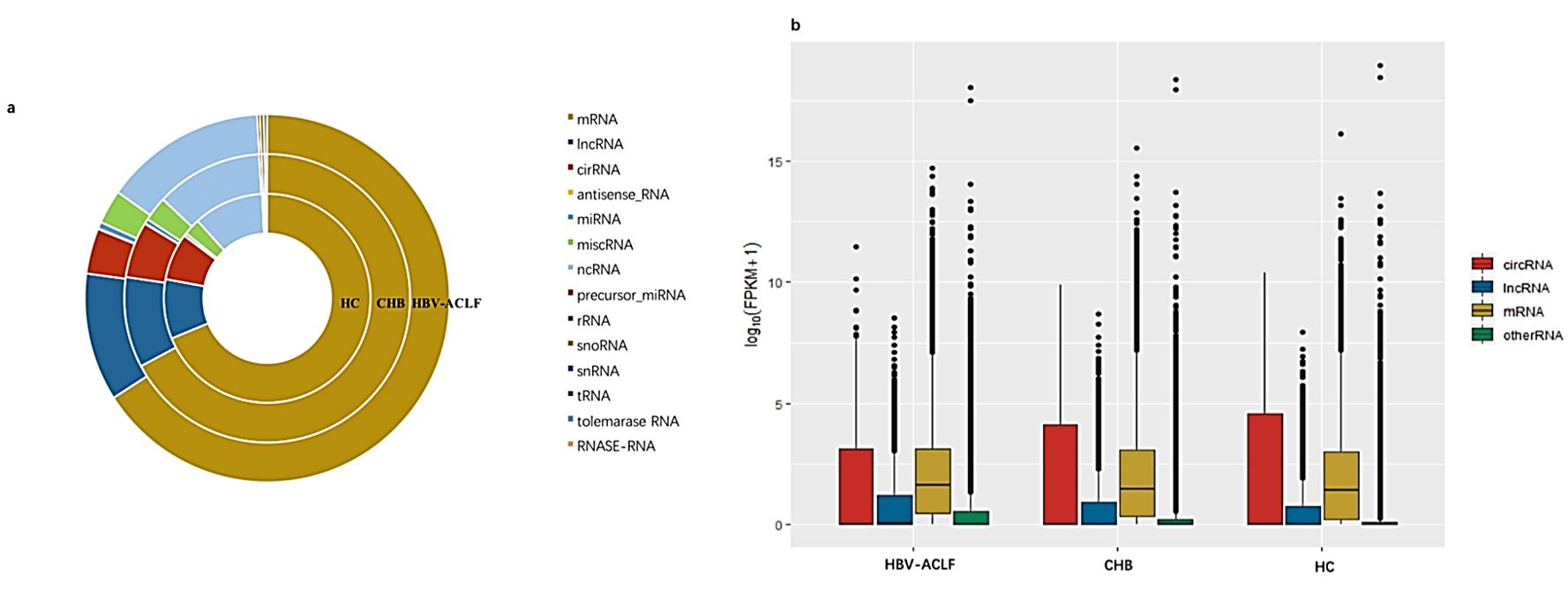 Figure 3. RNA expression profiles of serum exosomes from healthy individuals, CHB and HBV-ACLF. (Chen, J, et al., 2020)
Figure 3. RNA expression profiles of serum exosomes from healthy individuals, CHB and HBV-ACLF. (Chen, J, et al., 2020)
Our Services and Products
RNA sequencing can enhance understanding of the role of exosomes in a variety of physiological and pathological processes. By providing a non-invasive window into the molecular picture of cells and tissues, this powerful technology opens up new avenues for biomarker discovery, therapy development, and personalized medicine.
Creative Biostructure offers the integration of exosomal RNA sequencing with other cutting-edge technologies and advanced computational methods to help clients further unlock the potential of this exciting area of research. Please feel free to contact us for more information.
The products we offer are listed below.
| Cat No. | Product Name | Source |
| PNE-FB34 | PNExo™ Exosome-Blueberry | Exosome derived from Blueberry |
| PNE-FP93 | PNExo™ Exosome-Prickly Pear | Exosome derived from Prickly Pear |
| PNE-FLD10 | PNExo™ Exosome-Daffodil | Exosome derived from Daffodil |
| Exo-PDELN03 | HQExo™ Exosome-Onion | Exosome derived from Onion |
| Exo-PDELN04 | HQExo™ Exosome-Potato | Exosome derived from Potato |
| PNE-FLF63 | PNExo™ Exosome-Forget-me-not | Exosome derived from Forget-me-not |
| PNE-FLG70 | PNExo™ Exosome-Green Tea | Exosome derived from Green Tea |
| Exo-CH10 | HQExo™ Exosome-HCT116 | Exosome derived from human colorectal carcinoma cell line (HCT116 cell line) |
| Exo-CH11 | HQExo™ Exosome-K-562 | Exosome derived from human pleural effusion, leukemia chronic myelogenous (K-562 cell line) |
| Explore All Exosome Products | ||
References
- Elkommos-Zakhary M, et al. Exosome RNA Sequencing as a Tool in the Search for Cancer Biomarkers. Noncoding RNA. 2022. 8(6): 75.
- Lu J, et al. Small RNA sequencing analysis of exosomes derived from umbilical plasma in IUGR lambs. Commun Biol. 2023. 6(1): 943.
- Chen, J, et al. RNA Profiling Analysis of the Serum Exosomes Derived from Patients with Chronic Hepatitis and Acute-on-chronic Liver Failure Caused By HBV. Sci Rep. 2020. 1528.