Structural Research of Beta-sheet Pore-forming Toxins/Attack Complexes
Beta-sheet pore-forming toxins (β-PFTs) are a group of proteins that form pores in cell membranes through a series of conformational changes. β-strands are responsible for forming β-barrel and insert into the membrane. They are a widespread class of bacterial toxins and are known to cause various diseases. β-PFTs have been found in many bacteria, such as Listeria and Staphylococcus aureus. They are also involved in the virulence of some pathogens, making them an important target for the development of new treatments. Attack complexes are a subset of β-PFTs that work together as multi-subunit assemblies to create pores in host cell membranes, allowing for pathogenic organisms to enter and damage the host.
A recent study explores the structure of the translocational binary toxin complex CDTa-bound CDTb-pore from Clostridioides difficile, using cryo-electron microscopy (cryo-EM). The study provides insights into the mechanism of the binary toxin complex, observing that CDTb forms a barrel-shaped pore in the host cell membrane, with a central channel that allows for the translocation of CDTa. CDTa binds to the pore in a specific orientation, which is critical for its translocation into the host cell.
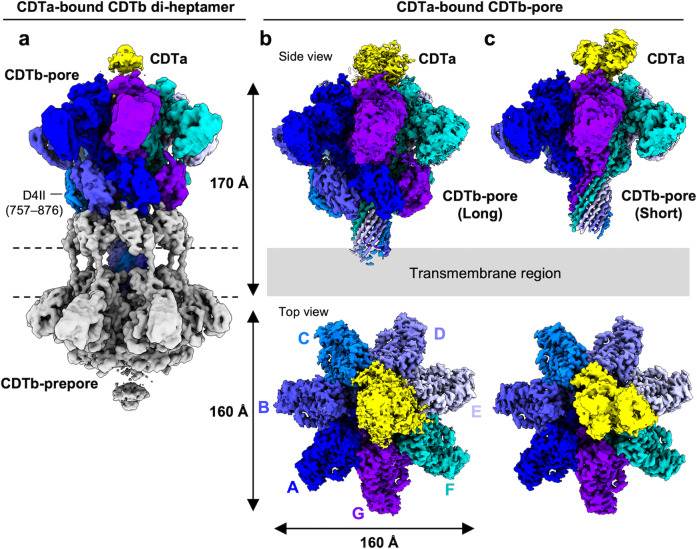 Figure 1. Cryo-EM density maps of CDTa-bound CDTb-pores. (Kawamoto A, et al., 2022)
Figure 1. Cryo-EM density maps of CDTa-bound CDTb-pores. (Kawamoto A, et al., 2022)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| α-hemolysin | Staphylococcus aureus | X-ray diffraction | 1.89 Å | 7AHL |
| M113F mutant of α-hemolysin (expressed in Staphylococcus aureus) | Staphylococcus aureus | X-ray diffraction | 2.10 Å | 3M2L |
| α-hemolysin (expressed in E. coli) | Staphylococcus aureus | X-ray diffraction | 2.30 Å | 3ANZ |
| α-hemolysin heptamer, C14-bound (expressed in E. coli) | Staphylococcus aureus | X-ray diffraction | 2.35 Å | 6U49 |
| γ-hemolysin composed of LukF and Hlg2 (expressed in E. coli) | Staphylococcus aureus | X-ray diffraction | 2.50 Å | 3B07 |
| γ-hemolysin prepore (expressed in E. coli) | Staphylococcus aureus | X-ray diffraction | 2.99 Å | 4P1Y |
| LukF component of γ-hemolysin (expressed in E. coli) | Staphylococcus aureus | X-ray diffraction | 1.90 Å | 1LKF |
| LUK prepore formed from LukF & LukS (expressed in E. coli) | Staphylococcus aureus | X-ray diffraction | 2.40 Å | 4P1X |
| LukGH octamer (expressed in E. coli) | Staphylococcus aureus | X-ray diffraction | 2.80 Å | 4TW1 |
| Leukocidin (LukFG) in complex with mouse CD11b I-domain (CD11b-I) (expressed in E. coli) | Staphylococcus aureus | X-ray diffraction | 2.29 Å | 6RHV |
| LukD, no ligand (expressed in E. coli) | Staphylococcus aureus | X-ray diffraction | 1.75 Å | 6U33 |
| leukotoxin LukE (expressed in E. coli) | Staphylococcus aureus | X-ray diffraction | 1.46 Å | 7P8T |
| Panton-Valentine leukocidin (PVL) with bound C14PC (expressed in E. coli) | Staphylococcus aureus | X-ray diffraction | 2.04 Å | 6U3Y |
| NetB Necrotic B-like enteritis toxin (expressed in E. coli) | Clostridium perfringens | X-ray diffraction | 3.90 Å | 4H56 |
| Perfringolysin O (PFO) protomer (expressed in E. coli) | Clostridium perfringens | X-ray diffraction | 2.20 Å | 1PFO |
| Anthrax Protective Antigen (PA) and Lethal Factor (LF) Prechannel Complex (expressed in E. coli) | Bacillus anthracis | X-ray diffraction | 3.10 Å | 3KWV |
| Anthrax protective antigen pore (expressed in E. coli) | Bacillus anthracis | Cryo-EM single particle analysis | 2.90 Å | 3J9C |
| Anthrax octamer prechannel bound to full-length edema factor (EF) (expressed in E. coli) | Bacillus anthracis | Cryo-EM single particle analysis | 3.30 Å | 6VRA |
| Anthrax protective antigen pore translating N-terminal of LF (expressed in E. coli) | Bacillus anthracis | Cryo-EM single particle analysis | 3.30 Å | 7KXR |
| Lymphocyte perforin monomer (expressed in Spodoptera frugiperda) | Mus musculus | X-ray diffraction | 2.75 Å | 3NSJ |
| Lymphocyte perforin pore (expressed in Spodoptera frugiperda) | Mus musculus | Cryo-EM single particle analysis | 4.00 Å | 7PAG |
| Macrophage-expressed gene 1 (MPEG1/Perforin-2), wild-type soluble pre-pore (expressed in Spodoptera frugiperda) | Homo sapiens | Cryo-EM single particle analysis | 3.49 Å | 6U23 |
| Macrophage-expressed gene 1 (MPEG1/Perforin-2), pore in ring form (expressed in HEK293 cells) | Mus musculus | Cryo-EM single particle analysis | 3.00 Å | 8A1D |
| Cytolysin pore-forming toxin (expressed in E. coli) | Vibrio cholerae | X-ray diffraction | 2.88 Å | 3O44 |
| Cytolysin pore-forming toxin protomer (expressed in E. coli) | Vibrio cholerae | X-ray diffraction | 2.30 Å | 1XEZ |
| Streptolysin O pore-forming toxin (expressed in E. coli) | Streptococcus pyogenes | X-ray diffraction | 2.10 Å | 4HSC |
| Aerolysin pore-forming toxin Y221G mutant (expressed in Aeromonas salmonicida) | Aeromonas hydrophila | X-ray diffraction | 2.88 Å | 3C0M |
| Monalysin pore-forming toxin, cleaved form (expressed in E. coli) | Pseudomonas entomophila | X-ray diffraction | 1.70 Å | 4MKO |
| poly-C9 component of the complement membrane attack Complex | Homo sapiens | Cryo-EM single particle analysis | 6.70 Å | 5FMW |
| poly-C9 component of the complement membrane attack Complex (expressed in Expi293 cells) | Homo sapiens | Cryo-EM single particle analysis | 3.90 Å | 6DLW |
| Membrane attack complex (MAC) in open conformation | Homo sapiens | Cryo-EM single particle analysis | 5.60 Å | 6H03 |
| 3C9-sMAC Complement membrane attack complex packaged for clearance | Homo sapiens | Cryo-EM single particle analysis | 3.54 Å | 7NYC |
| Monomeric C9 component of the complement membrane attack Complex (expressed in Expi293 cells) | Mus musculus | X-ray diffraction | 2.20 Å | 6CXO |
| Lysenin Pore complex (expressed in E. coli) | Eisenia fetida | Cryo-EM single particle analysis | 3.10 Å | 5GAQ |
| ILYml Cholesterol-dependent cytolysin, CD59-responsive (expressed in E. coli) | Streptococcus intermedius | X-ray diffraction | 2.89 Å | 5IMW |
| VLYml Cholesterol-dependent cytolysin, CD-59 responsive, bound to CD59D22A (expressed in E. coli) | Gardnerella vaginalis | X-ray diffraction | 2.40 Å | 5IMY |
| Pneumolysin (PLY) complex (expressed in E. coli) | Streptococcus pneumoniae | Cryo-EM single particle analysis | 4.50 Å | 5LY6 |
| Epsilon toxin (Etx) (expressed in E. coli) | Clostridium perfringens | Cryo-EM single particle analysis | 3.20 Å | 6RB9 |
| Binary iota toxin complex Ib pore (expressed in E. coli) | Clostridium perfringens | Cryo-EM single particle analysis | 2.90 Å | 6KLX |
| Binary toxin translocase CDTb in asymmetric tetradecamer conformation (expressed in E. coli) | Clostridioides difficile | Cryo-EM single particle analysis | 2.80 Å | 6UWR |
| Binary toxin translocase CDTb with CDTa long-stem pore (expressed in E. coli) | Clostridioides difficile | Cryo-EM single particle analysis | 2.64 Å | 7VNN |
Table 1. Structural Research of Beta-sheet Pore-forming Toxins/Attack Complexes.
Creative Biostructure is a leading provider of structural biology services that can assist researchers in advancing their understanding of these complex proteins. We offer a range of services that can help researchers determine the structure of β-PFTs and their attack complexes, including X-ray crystallography, NMR spectroscopy, and cryo-EM.
If you are interested in exploring the structural research of β-PFT or other membrane proteins, please don't hesitate to contact us. Our comprehensive structural biology services allow clients to leverage our expertise, beginning with protein expression and purification through to structure determination.
References
- Kawamoto A, et al. Cryo-EM structures of the translocational binary toxin complex CDTa-bound CDTb-pore from Clostridioides difficile. Nature Communications. 2022, 13(1): 6119.
- Trstenjak N, et al. Molecular mechanism of leukocidin GH–integrin CD11b/CD18 recognition and species specificity. Proceedings of the National Academy of Sciences. 2020, 117(1): 317-327.
- Lambey P, et al. Structural insights into recognition of chemokine receptors by Staphylococcus aureus leukotoxins. Elife. 2022, 11: e72555.
