Structural Research of Bacterial V-type ATPase
Bacterial V-type ATPases are molecular machines that play an essential role in the transport of ions across bacterial membranes. These enzymes are highly conserved across bacterial species and are involved in various biological processes, including nutrient acquisition, pH homeostasis, and virulence. Over the past decade, significant progress has been made in the structural analysis of bacterial V-type ATPases. Cryo-electron microscopy (Cryo-EM) has emerged as a powerful tool for the determination of high-resolution structures of these molecular machines. Several studies have reported the cryo-EM structures of V-ATPases from different bacterial species, including Thermus thermophilus, Enterococcus hirae, and Pyrococcus horikoshii.
The comprehensive study of the structural research of bacterial V-ATPases has unraveled the significant discovery of the rotary catalytic mechanism of these enzymes. It has been found that the intricate mechanism involves the rotation of a central rotor subunit within the enzyme, leading to the coupling of ATP hydrolysis to the transport of ions across the membrane. The domain structure of V-ATPases comprises of two functional domains, namely the V0 domain responsible for proton translocation, and the V1 domain responsible for ATP hydrolysis. The detailed analysis of these structures has facilitated the attainment of insights into the mechanism of energy transduction in biological systems, bringing us closer to understanding the fundamental principles that drive life.
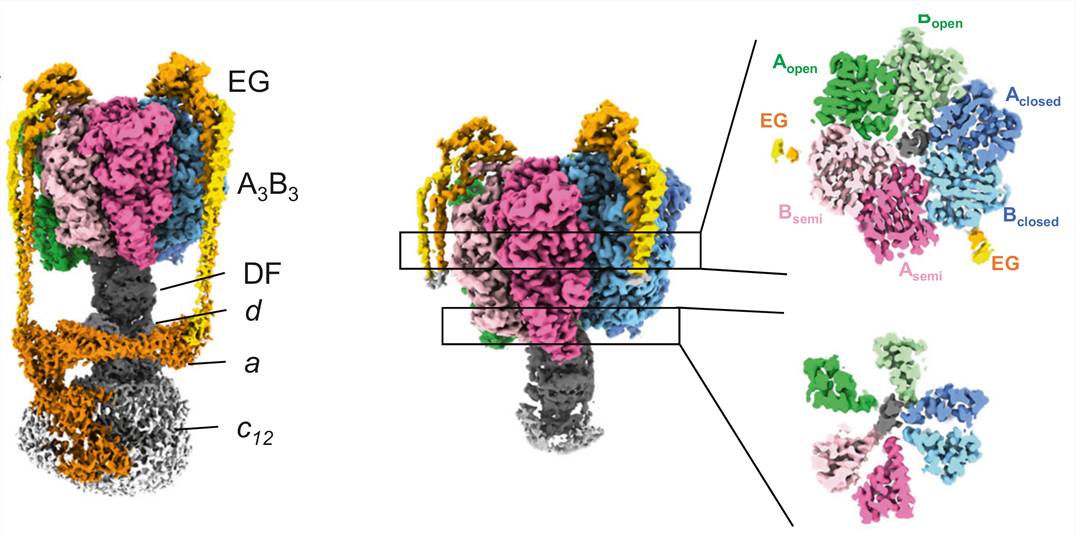 Figure 1. Cryo-EM density map for nucleotide-free V/A-ATPase. (Kishikawa J, et al., 2022)
Figure 1. Cryo-EM density map for nucleotide-free V/A-ATPase. (Kishikawa J, et al., 2022)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Rotor of V-type Na+-ATPase | Enterococcus hirae | X-ray diffraction | 2.10 Å | 2BL2 |
| Central axis (NtpD-NtpG) in the catalytic portion of V-type Na+-ATPase (expressed in Cell-free synthesis) | Enterococcus hirae | X-ray diffraction | 2.00 Å | 3AON |
| A3B3 Assembly of V-type Na+-ATPase (nucleotide-free, expressed in Cell-free synthesis) | Enterococcus hirae | X-ray diffraction | 2.80 Å | 3VR2 |
| 2 ADP-bound V1 complex (expressed in E. coli) | Enterococcus hirae | X-ray diffraction | 3.25 Å | 5KNB |
| Fitted atomic models of V-ATPase subunits into cryo-EM map | Thermus thermophilus | Cryo-EM single particle analysis | 9.70 Å | 3J0J |
| V1-ATPase Complex (V-ATPase soluble domain) with bound nucleotide | Thermus thermophilus | X-ray diffraction | 4.51 Å | 3A5C |
| A3B3 complex of V1-ATPase (expressed in E. coli) | Thermus thermophilus | X-ray diffraction | 2.80 Å | 3GQB |
| Peripheral stalk of H+-dependent V-ATP Synthase (expressed in E. coli) | Thermus thermophilus | X-ray diffraction | 3.10 Å | 3K5B |
| Peripheral stalk of H+-dependent V-ATP Synthase (expressed in E. coli) | Thermus thermophilus | X-ray diffraction | 2.25 Å | 3V6I |
| A3B3DF complex of V1-ATPase | Thermus thermophilus | X-ray diffraction | 3.90 Å | 3W3A |
| Complete V-type ATPase, rotation state 1 | Thermus thermophilus | Cryo-EM single particle analysis | 5.00 Å | 5Y5X |
| V/A-type ATPase/synthase in nanodiscs, rotational state 1 | Thermus thermophilus | Cryo-EM single particle analysis | 3.25 Å | 6QUM |
| V/A-ATPase in nanodiscs, soluble domain, including V1, d, two EG stalks, and N-terminal domain of a-subunit | Thermus thermophilus | Cryo-EM single particle analysis | 3.50 Å | 6LY8 |
| V1EG of V/A-ATPase, state1-1 | Thermus thermophilus | Cryo-EM single particle analysis | 3.10 Å | 7VAI |
| Peripheral stalk of H+-dependent V-ATP Synthase (expressed in E. coli) | Pyrococcus horikoshii | X-ray diffraction | 3.65 Å | 4DT0 |
Table 1. Structural Research of Bacterial V-type ATPase.
As a pioneering company that leads the structural analysis services, Creative Biostructure, boasts an exceptional team of experts who specialize in utilizing advanced techniques such as cryo-EM, X-ray crystallography, and NMR spectroscopy to gain a profound understanding of the structure and function of bacterial V-type ATPases from a plethora of bacterial species. Our clients can leverage our comprehensive structural analysis services, starting from protein expression and purification to structure determination. Our unwavering commitment to providing high-quality, accurate, and prompt structural analysis results remains unparalleled in the industry, a trait that continues to earn us a sterling reputation among our clients.
If you are interested in exploring the structural research of bacterial V-type ATPases and want to learn more about our services, please don't hesitate to contact us. Our team is always available to discuss your research needs and offer the best possible solutions for your project. Let us work together to advance scientific knowledge in this field and contribute to the betterment of life.
References
- Kishikawa J, et al. Structural snapshots of V/A-ATPase reveal the rotary catalytic mechanism of rotary ATPases. Nature Communications. 2022, 13(1): 1213.
- Kishikawa J, et al. Mechanical inhibition of isolated Vo from V/A-ATPase for proton conductance. Elife. 2020, 9: e56862.