Structural Research of Electron Transport Chain Complex III
Electron transport chain (ETC) is an essential metabolic process that occurs in the inner mitochondrial membrane and is crucial for the generation of ATP through oxidative phosphorylation. Complex III, also known as cytochrome bc1 complex, is a crucial component of ETC, which plays a vital role in transferring electrons from ubiquinol to cytochrome c and translocating protons across the membrane. Recent advancements in structural analysis techniques have enabled researchers to gain deeper insights into the structure and mechanism of complex III.
Researchers have made significant strides in the field of structural biology, with cryo-electron microscopy (cryo-EM) emerging as a leading technique for determining high-resolution structures of biological macromolecules. Cryo-EM enables the visualization of protein structures in near-native conditions, providing detailed information on their function and interactions with other biomolecules.
A recent study used cryo-EM to determine the structures of Candida albicans complex III homodimer (CIII2), revealing endogenous ubiquinone and visualizing the continuum of Rieske head domain conformations. The analysis of these conformations did not indicate cooperativity in the Rieske head domain position or ligand binding in the two CIIIs of the CIII2 dimer. The study also used cryo-EM with the indazole derivative Inz-5, which inhibits fungal CIII2 and is fungicidal when administered with fungistatic azole drugs, to show that Inz-5 inhibition alters the equilibrium of Rieske head domain positions. These findings provide valuable insights into the mechanism of action of CIII2 and the effects of Inz-5 inhibition on the Rieske head domain.
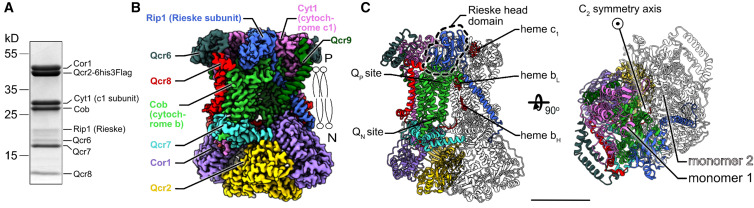 Figure 1. Overall structure of Candida albicans complex III. (Di Trani J M, et al., 2022)
Figure 1. Overall structure of Candida albicans complex III. (Di Trani J M, et al., 2022)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Cytochrome bc1 | Bos taurus | X-ray diffraction | 2.70 Å | 1QCR |
| Cytochrome bc1 | Bos taurus | X-ray diffraction | 3.00 Å | 1BGY |
| Cytochrome bc1 | Bos taurus | X-ray diffraction | 2.26 Å | 2FYU |
| Cytochrome bc1 | Bos taurus | X-ray diffraction | 2.40 Å | 1NTM |
| Cytochrome bc1 | Bos taurus | X-ray diffraction | 2.60 Å | 1SQX |
| Cytochrome bc1 | Bos taurus | X-ray diffraction | 2.10 Å | 1PPJ |
| Cytochrome bc1 in complex with azoxystrobin (expressed in Bos taurus) | Bos taurus | X-ray diffraction | 2.80 Å | 6NHG |
| Cytochrome bc1 in complex with tetrahydro-quinolone inhibitor JAG021 | Bos taurus | X-ray diffraction | 3.50 Å | 6XVF |
| Cytochrome bc1 in complex with inhibitor CK-2-67 | Bos taurus | X-ray diffraction | 3.20 Å | 7R3V |
| Cytochrome bc1 | Gallus gallus | X-ray diffraction | 3.16 Å | 1BCC |
| Cytochrome bc1 (expressed in E. coli) | Saccharomyces cerevisiae | X-ray diffraction | 2.30 Å | 1EZV |
| Cytochrome bc1 (expressed in E. coli) | Saccharomyces cerevisiae | X-ray diffraction | 2.30 Å | 1KB9 |
| Cytochrome bc1 (expressed in E. coli) | Saccharomyces cerevisiae | X-ray diffraction | 2.50 Å | 1P84 |
| Cytochrome bc1 (expressed in E. coli) | Saccharomyces cerevisiae | X-ray diffraction | 2.30 Å | 2IBZ |
| Cytochrome bc1 (expressed in E. coli) | Saccharomyces cerevisiae | X-ray diffraction | 1.90 Å | 3CX5 |
| Cytochrome bc1 (expressed in Cereibacter sphaeroides) | Cereibacter sphaeroides | X-ray diffraction | 3.20 Å | 2FYN |
| Cytochrome bc1 with azoxystrobin | Cereibacter sphaeroides | X-ray diffraction | 3.00 Å | 6NHH |
| Cytochrome bc1 (expressed in Rhodobacter capsulatus) | Rhodobacter capsulatus | X-ray diffraction | 3.51 Å | 1ZRT |
| quinol:cytochrome c/HiPIP oxidoreductase (alternative complex III) | Rhodothermus marinus DSM 4252 | Cryo-EM single particle analysis | 3.87 Å | 6F0K |
| Alternative complex III (ACIII) in SMA nanodiscs | Flavobacterium johnsoniae UW101 | Cryo-EM single particle analysis | 3.40 Å | 6BTM |
| Alternative complex III (ACIII), dithionite reduced | Roseiflexus castenholzii DSM 13941 | Cryo-EM single particle analysis | 3.50 Å | 6LOE |
| Cytochrome bc1 apo structure | Aquifex aeolicus VF5 | Cryo-EM single particle analysis | 3.30 Å | 6KLS |
| Complex III2 (Cytochrome bc1 homodimer), inhibitor free | Candida albicans SC5314 | Cryo-EM single particle analysis | 3.00 Å | 7RJA |
| Complex III2 (Cytochrome bc1 homodimer), combined datasets, consensus refinement | Yarrowia lipolytica | Cryo-EM single particle analysis | 2.00 Å | 8AB6 |
Table 1. Structural Research of Electron Transport Chain Complex III.
At Creative Biostructure, we offer high-quality structural analysis services for electron transport chain complex III and other biological macromolecules. Our team of highly skilled scientists has extensive experience using advanced technologies such as cryo-EM, X-ray crystallography, and nuclear magnetic resonance (NMR) spectroscopy to provide comprehensive structural information about the molecules under study. We offer a range of services including protein crystallization, structure determination, refinement, and analysis, and can also customize our solutions to meet the specific needs of our clients. Our expertise in membrane protein sample preparation and experimental conditions optimization enables us to provide tailored services that meet the unique requirements of each research project. With our cutting-edge facilities and personalized approach, we can help researchers gain a deeper understanding of the complex molecular mechanisms of biological systems, including complex III. Our team is dedicated to providing exceptional customer service and support to our clients, ensuring their satisfaction. Contact us to learn more about how we can assist you in your research endeavors.
References
- Esser L, et al. Crystal structure of bacterial cytochrome bc1 in complex with azoxystrobin reveals a conformational switch of the Rieske iron–sulfur protein subunit. Journal of Biological Chemistry. 2019, 294(32): 12007-12019.
- Di Trani J M, et al. Rieske head domain dynamics and indazole-derivative inhibition of Candida albicans complex III. Structure. 2022, 30(1): 129-138. e4.
- Wieferig J P, Kühlbrandt W. Analysis of the conformational heterogeneity of the Rieske iron–sulfur protein in complex III2 by cryo-EM. IUCrJ. 2023, 10(1).