Structural Research of G Protein-coupled Receptors (GPCRs) Class B2
G Protein-coupled Receptors (GPCRs) are a superfamily of cell membrane receptors that play a crucial role in cellular signaling and response to various extracellular stimuli. Among the different classes of GPCRs, class B2, also known as the adhesion GPCR family, stands as one of the least understood groups. The adhesion GPCR family exhibits a distinctive molecular structure that distinguishes them from other GPCR classes. GPCRs class B2 possess an extended N-terminal portion comprising diverse adhesion domains. Furthermore, they contain a well-conserved GPCR autoproteolysis-inducing (GAIN) domain immediately preceding the first transmembrane helix.
A defining characteristic of the aGPCR family is their autoproteolytic cleavage at a highly conserved GPCR proteolysis site (GPS) located within the GAIN domain. This intriguing feature has been proposed to be critical for the maturation and proper functioning of these receptors. Upon cleavage, the receptors generate two noncovalently associated fragments: an N-terminal fragment (NTF) consisting mostly of the extracellular domain, and a C-terminal fragment (CTF) containing a portion of the proteolysed GAIN domain and the transmembrane domain (TMD).
Recent studies further present research findings on two specific aGPCRs, ADGRD1 and ADGRF1. The study reveals that the stalk region preceding the first transmembrane helix acts as the tethered agonist by interacting extensively with the transmembrane domain. This stalk-transmembrane domain interaction pattern is shared among members of the aGPCR family. Interestingly, the study also indicates a cleavage-independent manner of receptor activation, supported by the structure of autoproteolysis-deficient ADGRF1. The activation process involves a cascade of inter-helix interaction cores, which are conserved in position but show sequence variability in ADGRD1 and ADGRF1. Additionally, the intracellular region of ADGRF1 contains a specific lipid-binding site, which is functionally significant and may be involved in recognizing the endogenous ADGRF1 ligand synaptamide.
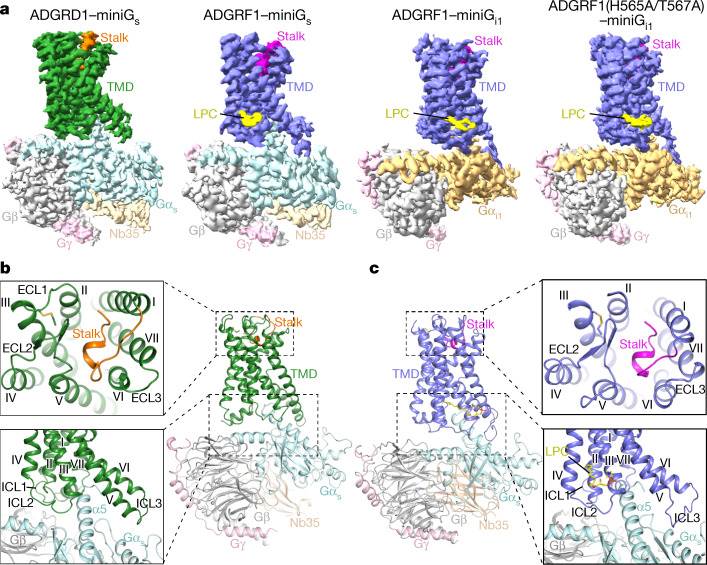 Figure 1. Overall structures of G protein-bound ADGRD1 and ADGRF1. (Qi X, et al., 2022)
Figure 1. Overall structures of G protein-bound ADGRD1 and ADGRF1. (Qi X, et al., 2022)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Adhesion receptor GPR133 (ADGRD1)-miniGs complex (expressed in Spodoptera frugiperda) | Homo sapiens | Cryo-EM single particle analysis | 2.80 Å | 7WU2 |
| Adhesion receptor GPR110 (ADGRF1)-miniGs complex (expressed in Spodoptera frugiperda) | Homo sapiens | Cryo-EM single particle analysis | 3.10 Å | 7WU3 |
| Adhesion receptor GPR110 (ADGRF1) - Gq complex (expressed in Spodoptera frugiperda) | Homo sapiens | Cryo-EM single particle analysis | 2.85 Å | 7WXU |
| Adhesion receptor GPR56 (ADGRG1)-miniG13 complex with bound tethered agonist (expressed in Spodoptera frugiperda) | Homo sapiens | Cryo-EM single particle analysis | 2.70 Å | 7SF8 |
| Adhesion receptor GPR97 (ADGRG3)-Go complex with bound beclomethasone (expressed in Spodoptera frugiperda) | Homo sapiens | Cryo-EM single particle analysis | 3.10 Å | 7D76 |
| Adhesion receptor LPHN3 (ADGRL3)-miniG13 complex with bound tethered agonist (expressed in Spodoptera frugiperda) | Homo sapiens | Cryo-EM single particle analysis | 2.90 Å | 7SF7 |
| Adhesion receptor LPHN3 (ADGRL3)-Gq complex (expressed in Insect BA phytoplasma) | Homo sapiens | Cryo-EM single particle analysis | 2.83 Å | 7WY5 |
Table 1. Structural research of class B2 GPCRs.
At Creative Biostructure, we understand that each research project is unique. We offer personalized solutions and flexible collaboration opportunities to meet our clients' specific needs. Whether it's a basic structural study or a comprehensive investigation into aGPCRs' functional aspects, we are dedicated to delivering results that drive scientific progress. We are well-equipped with state-of-the-art technologies and cutting-edge instrumentation, allowing us to provide high-resolution structural analysis services with unmatched precision. Our capabilities include including X-ray crystallography, cryo-electron microscopy (cryo-EM), and nuclear magnetic resonance (NMR) spectroscopy. Our team consists of highly experienced scientists, structural biologists, and researchers who work synergistically to address intricate biological inquiries. Contact us to explore how our advanced resources can elevate your research endeavors and bring you closer to accomplishing your scientific objectives.
References
- Qu X, et al. Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1. Nature. 2022, 604(7907): 779-785.
- Zhu X, et al. Structural basis of adhesion GPCR GPR110 activation by stalk peptide and G-proteins coupling. Nature Communications. 2022, 13(1): 5513.