Structural Research of Type III Secretion Systems
The type III secretion system (T3SS) is a large transmembrane protein mechanism that delivers effector proteins from the cytoplasm of pathogenic bacteria to eukaryotic host cells via the injectome, which is essential for the motility and virulence of many major Gram-negative bacterial pathogens. Research has shown that the T3SS is an attractive target for the discovery or design of novel anti-infective drug and vaccine approaches, and understanding the structure of the T3SS is essential for developing therapeutics targeting this system.
Structure of the T3SS injectosome
The core conserved protein of T3SS forms a transmembrane syringe-like structure. The injectosome spans the bacterial inner and outer membranes and has extracellular filamentous appendages that extend into the target host cell. It presents effector proteins to the cytoplasmic sorting platform complex in a secreted state, after which the effectors are sorted and loaded into the output apparatus (EA) subcomplex located inside the membrane-bound matrix. Extending from the EA is a long helical needle filament, and the tip complex contacts the host cell membrane through the assembly of translocation subpores. The matrix and the needle filament, collectively called the needle complex, act as a continuous conduit for effector protein translocation from the prokaryotic cytoplasm to the host cell cytoplasm.
Advances in T3SS structure research
Researchers have recently obtained several high-resolution cryo-electron microscopy (cryo-EM) structures by single-particle analysis of needle complexes. In T3SS, the N-terminal domains N0 and N1 form a connector, and N3, together with trypticin and the S-structural domain, forms an outer membrane (OM) ring. The OM ring is embedded in the outer membrane, forming a pentameric (C15 symmetric) assembly that assembles into a homo-oligomeric β-barrel. The inner wall forms a ring of approximately 70 Å diameter. At the distal end, the outer four β-strands of each monomer form a sub-structural domain with a hydrophobic L-shaped groove. The proximal opening of the OM ring is lined with an N3 structural domain containing a conserved ring-building motif (RBM).
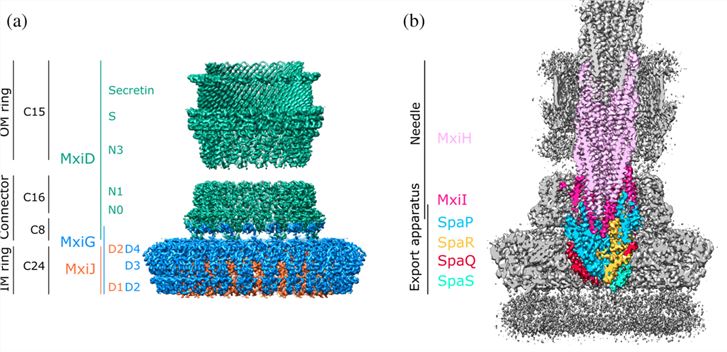 Figure 1. Structural overview of the type 3 secretion system needle complex. (Flacht L, et al., 2023)
Figure 1. Structural overview of the type 3 secretion system needle complex. (Flacht L, et al., 2023)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Type III secretion system protein | Salmonella enterica subsp. enterica serovar Typhimurium | X-ray diffraction | 1.858 Å | 4G2S |
| Type 3 secretion system export apparatus core, inner rod, and needle | Shigella flexneri | Cryo-EM single particle analysis | 4.05 Å | 8AXK |
| Inner membrane ring and secretin N0 N1 domains of the type 3 secretion system | Shigella flexneri | Cryo-EM single particle analysis | 3.34 Å | 8AXN |
| Export apparatus core and inner rod of the type 3 secretion system | Shigella flexneri | Cryo-EM single particle analysis | 5.11 Å | 6RWY |
| Type three secretion system protein | Escherichia coli | X-ray diffraction | 1.9 Å | 2OVS |
| Outer membrane secretin pore of the type 3 secretion system | Shigella flexneri | Cryo-EM single particle analysis | 3.42 Å | 8AXL |
| MxiD N0 N1 and MxiG C-terminal domains of the type 3 secretion system | Shigella flexneri | Cryo-EM single particle analysis | 3.86 Å | 6RWK |
| Periplasmic inner membrane ring of the type 3 secretion system | Shigella flexneri | Cryo-EM single particle analysis | 3.55 Å | 6RWX |
| Assembled ATPase EscN in complex with its central stalk EscO from the type III secretion system | Escherichia coli O127:H6 str. E2348/69 | Cryo-EM single particle analysis | 3.29 Å | 6NJP |
| Secretin pilot protein from the type III secretion system (T3SS) | Pseudomonas aeruginosa PAO1 | X-ray diffraction | 1.81 Å | 2YJL |
| T3SS basal body | Salmonella enterica subsp. enterica serovar Typhimurium | Cryo-EM single particle analysis | 11.7 Å | 3J1W |
| Highly conserved ATPase from type III secretion system | Shigella flexneri | X-ray diffraction | 2.5 Å | 5YBH |
| Assembled ATPase EscN from the type III secretion system | Escherichia coli O127:H6 str. E2348/69 | Cryo-EM single particle analysis | 3.34 Å | 6NJO |
| T3SS basal body component YscD | Yersinia enterocolitica | X-ray diffraction | 1.4 Å | 4ALZ |
| Type III secretion system ATPase InvC | Salmonella enterica subsp. enterica serovar Typhimurium str. SL1344 | X-ray diffraction | 2.049 Å | 6RAE |
| Type III Secretion system protein OrgC | Salmonella enterica subsp. enterica | SOLUTION NMR | / | 6CJD |
| Prototypical ATPase from the type III secretion system | Escherichia coli O127:H6 str. E2348/69 | X-ray diffraction | 2.25 Å | 2OBM |
| T3SS tip protein LcrV (G28-D322, C273S) | Yersinia pestis | X-ray diffraction | 1.65 Å | 4JBU |
| The cytoplasmic domain of the T3SS sorting platform protein PscD | Pseudomonas aeruginosa | X-ray diffraction | 1.45 Å | 6UID |
| PscU C-terminal domain | Pseudomonas aeruginosa PAO1 | X-ray diffraction | 2.9 Å | 5CUL |
| Type III secretion injectisome EspA filament | Escherichia coli O127:H6 str. E2348/69 | Cryo-EM single particle analysis | 3.56 Å | 7K7K |
| Apo-state type 3 secretion system export apparatus complex | Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 | Cryo-EM single particle analysis | 3.6 Å | 7AGX |
| The C-terminal domain of the type III effector IpaH | Shigella flexneri 2a str. 301 | X-ray diffraction | 2.65 Å | 3CKD |
| The nonameric EscV cytosolic domain from the type III secretion system | Escherichia coli O127:H6 str. E2348/69 | Cryo-EM single particle analysis | 4.7 Å | 7K08 |
Table 1. Structural research of the type III secretion systems.
As a leading company in structural biology, Creative Biostructure offers a range of services dedicated to the research of type III secretion systems (T3SS) and other biomolecules. Drawing on a wealth of expertise and cutting-edge technology, we offer a comprehensive range of structural analysis services to both academia and the biotechnology industry.
Our team of expert scientists utilizes cutting-edge techniques such as X-ray crystallography, cryo-electron microscopy (cryo-EM), and NMR spectroscopy to acquire high-resolution structural data. Contact us to learn how our advanced capabilities can enhance your research and propel you closer to achieving your scientific goals.
References
- Flacht L, et al. Integrative structural analysis of the type III secretion system needle complex from Shigella flexneri. Protein Sci. 2023. 32(4): e4595.
- Miletic S, et al. Substrate-engaged type III secretion system structures reveal gating mechanism for unfolded protein translocation. Nat Commun. 2021. 12(1): 1546.
- Notti RQ, Stebbins CE. The Structure and Function of Type III Secretion Systems. Microbiol Spectr. 2016. 4(1).
