Structural Research of Acyltransferases
Acyltransferases (ATases) are a fundamental class of enzymes whose main function is to catalyze the acylation and deacylation of proteins. Acyltransferases are a large family of multifunctional proteins, and different acyltransferases play important roles in the genetics, gene expression, substance metabolism, and signaling of organisms.
The overall structure of an acyltransferase is usually compact and spherical or rod-shaped. It usually contains a catalytic structural domain with acyl transfer activity and one or more regulatory structural domains. The catalytic domain usually contains a set of conserved amino acid residues that are essential for catalysis. Lipid A acyltransferase (LpxM) and intact membrane acyltransferase (PatA) are two common ATases.
Lysophospholipid acyltransferase (LPLAT) proteins are required for many essential biological activities involving acyl chain transfer. One of the LPLATs, LpxM, is required for the biosynthesis of lipid A. The spatial structure of LpxM is spherical and is bisected by a large seven-stranded β-fold plate. The predicted transmembrane structural domain consists of a single α-helix that protrudes from the spherical structural domain and forms a substantial crystal contact with LpxM in the adjacent asymmetric unit. Electrostatic surface visualization of the LABLAT LpxM (Ab LpxM) structure from the pathogenic bacterium Acinetobacter baumannii reveals a very large hydrophobic pocket that binds a co-purified n-dodecyl-β- d-maltoside (DDM) molecule. Elucidation of the bacterial LABLAT structure may help to identify compounds that interfere with lipid A biosynthesis, leading to membrane leakage, thereby reducing virulence and enhancing the uptake of bactericidal compounds.
PatA is an essential acyltransferase involved in the biosynthesis of phosphatidylinositol mannosides (PIM), a key structural component of Mycobacterium tuberculosis, and virulence factors. The tertiary structure of PatA consists of the N-terminal transmembrane domain, the N-terminal cytoplasmic domain, and the C-terminal cytoplasmic region. the C-terminal cytoplasmic domain consists mainly of an α-helix and a β-fold, which is connected to the N-terminal cytoplasmic region to form the complete cytoplasmic structure. One of the β-folds of this structural domain is located at the C-terminus and contains key residues of the catalytic mechanism that catalyzes the acyl transfer reaction.
The electrostatic potential of the PatA surface reveals a clear solvent-exposed region bordering the main groove, which contains several hydrophobic and aromatic residues, scattered with positively charged residues. Specifically, this region contains the amphiphilic helices α3, α4, and α8, as well as the linker ring β2-α8. In contrast, the other side of PatA shows a negatively charged surface, which would produce significant electrostatic repulsion with the anionic phospholipid bilayer.
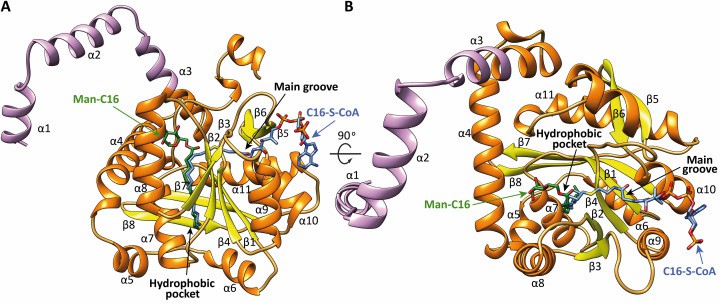 Figure 1. Two views of the x-ray crystal structures of PatA. (Anso I, et al., 2021)
Figure 1. Two views of the x-ray crystal structures of PatA. (Anso I, et al., 2021)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| LpxM lipid A acyltransferase (expressed in E. coli) | Acinetobacter baumannii NIPH 410 | X-ray diffraction | 1.99 Å | 5KN7 |
| PatA membrane-associated acyltransferase | Mycolicibacterium smegmatis MC2 155 | X-ray diffraction | 3.67 Å | 7OJT |
Table 1. Structural Research of Acyltransferases.
X-ray crystallography is a powerful technique widely used in structural biology. The principle of this technology is to obtain highly detailed and accurate protein structure images by irradiating an X-ray beam onto a molecular crystal and measuring the resulting diffraction patterns. Very suitable for analyzing the three-dimensional structure of membrane proteins.
If you need to ensure that the structure and function of proteins are accurately and comprehensively analyzed and understood, Creative Biostructure's X-ray crystallography service is your gold-standard choice. We use state-of-the-art high-intensity X-ray and special detectors to provide high-resolution images of protein crystal structures. If you are interested in learning more about our protein structural analysis services, please contact us for more information.
References
- Dovala D, et al. Structure-guided enzymology of the lipid a acyltransferase LpxM reveals a dual activity mechanism. Proceedings of the National Academy of Sciences, 2016, 113(41).
- Anso I, et al. Molecular ruler mechanism and interfacial catalysis of the integral membrane acyltransferase pata. Science Advances, 2021, 7(42).