Structural Research of Type IV Secretion Systems
The type IV secretion system (T4SS) is found in Gram-negative and Gram-positive bacteria and is a functionally diverse translocation superfamily. It typically translocates DNA and effector proteins into eukaryotic target cells by relying on direct cell-to-cell contact. In the past, X-ray crystallography and cryo-electron microscopy have achieved essential milestones in research, gradually revealing the shared structural features of T4SS and how structural variations confer specific functional diversity.
Advances in T4SS research
T4SS consists of two major subfamilies, splicing systems and effector transposons. Splice systems mediate DNA transfer between bacteria and are responsible for rapidly transmitting antibiotic-resistance genes and virulence determinants. Many Gram-negative bacterial pathogens use effector transposons to deliver virulence macromolecules into prokaryotic or eukaryotic cells to regulate physiological processes during infection. Recently, researchers have made great progress in defining the structure of T4SS units and components.
T4SS structural analysis
Researchers demonstrated the cryo-electron microscopy (cryo-EM) structure of the T4SS complex from the R388 plasmid at 2.8 Å resolution. T4SS is found to contain four sub-complexes consisting of 12 proteins (VirB1-VirB11 and VirD4), the outer membrane core complex (OMCC), the stem, the arch, and the inner membrane complex (IMC). In the IMC, six dimers of VirB4 are clustered together, where the interface between two adjacent VirB4 central subunits is distributed over the N- and C-terminal structural domains. The stem is a central conical assembly consisting of VirB6 pentamers and VirB5 pentamers. The arch consists of a hexamer of the homotrimeric unit of VirB8 peri, forming a ring around the stem. The OMCC consists of an O-layer embedded in the outer membrane and an I-layer located below the periplasm.
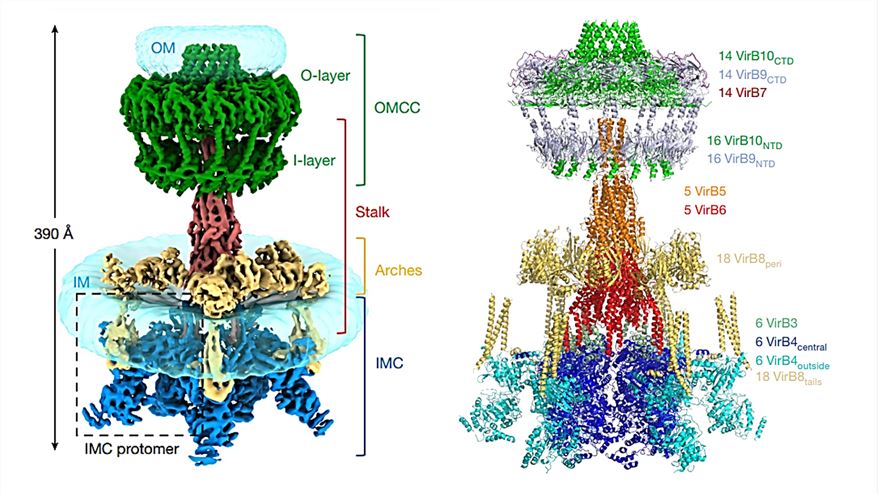 Figure 1. Composite electron density map (left) and overall model (right) of R388 T4SS. (Macé K, et al., 2022)
Figure 1. Composite electron density map (left) and overall model (right) of R388 T4SS. (Macé K, et al., 2022)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| T4SS protein CagL | Helicobacter pylori 26695 | X-ray diffraction | 3.25 Å | 3ZCJ |
| Dot T4SS OMC | Legionella pneumophila | Cryo-EM single particle analysis | 3.5 Å | 6X62 |
| Dot T4SS PR | Legionella pneumophila | Cryo-EM single particle analysis | 3.7 Å | 6X64 |
| Dot/Icm T4SS | Legionella pneumophila | Cryo-EM single particle analysis | 3.7 Å | 6X65 |
| dDot T4SS OMC | Legionella pneumophila | Cryo-EM single particle analysis | 4.2 Å | 6X66 |
| CagL-K74 | Helicobacter pylori | X-ray diffraction | 2.791 Å | 4YVM |
| Stalk complex structure (TrwJ/VirB5-TrwI/VirB6) | Escherichia coli | Cryo-EM single particle analysis | 3.7 Å | 7O3V |
| VirB8 domain of PrgL | Enterococcus faecalis | X-ray diffraction | 1.735 Å | 7AED |
| Dot/Icm T4SS PR | Legionella pneumophila | Cryo-EM single particle analysis | 2.8 Å | 7MUE |
| Dot/Icm T4SS C1 | Legionella pneumophila | Cryo-EM single particle analysis | 3.8 Å | 7MUC |
| T4SS Protein CagL | Helicobacter pylori 26695 | X-ray diffraction | 2.15 Å | 4CII |
| The BID Domain of Bep6 | Bartonella rochalimae ATCC BAA-1498 | X-ray diffraction | 2.1 Å | 4YK1 |
| CagV, a VirB8 homolog of T4SS | Helicobacter pylori 26695 | X-ray diffraction | 1.922 Å | 6IQT |
| T4SS protein CagL | Helicobacter pylori 26695 | X-ray diffraction | 2.201 Å | 3ZCI |
| CagL | Helicobacter pylori 26695 | X-ray diffraction | 2.3 Å | 4X5U |
| CagX from T4SS | Helicobacter pylori | Cryo-EM single particle analysis | 3.8 Å | 6OGE |
| CagT from T4SS | Helicobacter pylori | Cryo-EM single particle analysis | 3.8 Å | 6OEE |
| CagY from T4SS | Helicobacter pylori | Cryo-EM single particle analysis | 3.8 Å | 6ODI |
| PolyAla model of OMCC I-layer | Helicobacter pylori | Cryo-EM single particle analysis | 3.8 Å | 6OEH |
| PolyAla model of the PRC from the type 4 secretion system | Helicobacter pylori | Cryo-EM single particle analysis | 3.5 Å | 6ODJ |
| PolyAla model of the O-layer from the type 4 secretion system | Helicobacter pylori | Cryo-EM single particle analysis | 3.8 Å | 6OEF |
| TraC | Escherichia coli | X-ray diffraction | 3 Å | 1R8I |
| VirB9 C-terminal domain in complex with VirB7 N-terminal domain from T4SS | Xanthomonas citri pv. citri str. 306 | SOLUTION NMR | / | 2N01 |
| Cagbeta with CagZ | Helicobacter pylori 26695 | X-ray diffraction | 2.1 Å | 6JHO |
| C13 reconstruction of outer membrane core complex (OMCC) of type IV secretion system (T4SS) | Salmonella enterica subsp. enterica serovar Typhi | Cryo-EM single particle analysis | 3.31 Å | 7SPB |
| C17 reconstruction of outer membrane core complex (OMCC) of type IV secretion system (T4SS) | Salmonella enterica subsp. enterica serovar Typhi | Cryo-EM single particle analysis | 2.95 Å | 7SPC |
Table 1. Structural research of the type IV secretion systems.
Creative Biostructure is a leading biological company offering a range of protein structure analysis services including X-ray crystallography, cryo-electron microscopy (cryo-EM), and NMR spectroscopy. Our team of experienced structural biologists has extensive expertise in characterizing membrane protein structures and can provide high-quality structural information for challenging targets.
In addition, we provide related services including protein expression and purification, biophysical characterization, and computational modeling to facilitate new advances in researchers' studies of the structure and action mechanisms of the T4SS. If you are interested in the structural analysis of T4SS or other biomolecules, please feel free to contact us. We are committed to providing high-quality data and support to our clients and look forward to working with you on your structural biology projects.
References
- Macé K, et al. Cryo-EM structure of a type IV secretion system. Nature. 2022. 607(7917): 191-196.
- Liu X, et al. Structure of a type IV secretion system core complex encoded by multi-drug resistance F plasmids. Nat Commun. 2022. 13(1): 379.
- Sheedlo MJ, et al. Molecular architecture of bacterial type IV secretion systems. PLoS Pathog. 2022. 18(8): e1010720.
- Christie PJ, et al. Mechanism and structure of the bacterial type IV secretion systems. Biochim Biophys Acta. 2014. 1843(8): 1578-1591.
