Protein-Protein Interation
Proteins function not only as individual units but also interact with other proteins to facilitate complex biological functions. These interactions, designated as protein-protein interactions (PPIs), are pivotal to the structural and functional organization of cells. PPIs involve physical contacts between two or more protein molecules as a consequence of biochemical processes and electrostatic forces. These interactions can be transient or construct stable complexes. PPIs are central to numerous cellular processes including signal transduction, structural support, cell transport, and regulation of cellular activities.
At Creative Biostructure, we are dedicated to delivering top-tier PPI assay services and exceptional outcome solutions to advance your research and pharmaceutical endeavors.
Types of Protein-Protein Interactions
PPIs can be broadly categorized based on their different features:
Homo-Oligomers vs. Hetero-Oligomers
Homo-oligomers are protein complexes that are formed by the combination of identical protein subunits. These subunits interact with one another to assembly a symmetrical structure. Homo-oligomers frequently display symmetrical arrangements, including dimers, trimers, tetramers, and so forth. It has been observed that the formation of homo-oligomers increases the stability of the individual protein subunits. Homo-oligomerization has the potential to regulate the functional activity of the protein, such as increasing enzymatic activity or facilitating regulatory functions. Notable examples include hexokinase 1 and collagen.
In contrast, hetero-oligomers are protein complexes that are formed through the assembly of non-identical subunits. The subunits may be different proteins or different isoforms of a single protein. The structural and functional diversity exhibited by hetero-oligomers allows for the implementation of intricate regulatory mechanisms. The interaction between different subunits can confer specificity to the complex, thereby enabling precise cellular functions. The formation of hetero-oligomers can facilitate the assembly of different functional domains, thereby creating a diverse complex with combined activities. Two notable examples are the ribosome and the DNA polymerase holoenzyme.
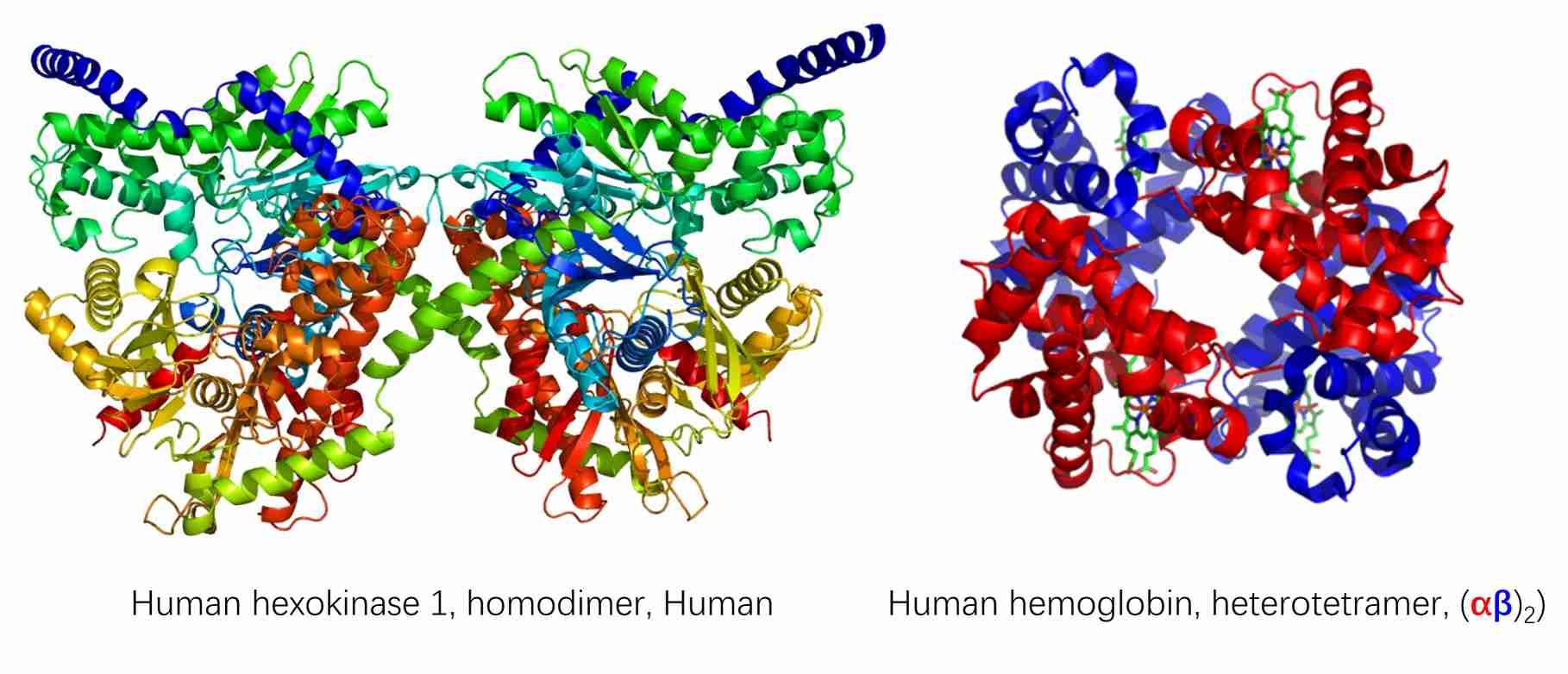 Figure 1. Example of homo-oligomer (hexokinase 1) and hetero-oligomer (hemoglobin) (Protein Data Bank).
Figure 1. Example of homo-oligomer (hexokinase 1) and hetero-oligomer (hemoglobin) (Protein Data Bank).
Obligate vs. Non-Obligate
In obligate interactions, the protein partners are unable to function independently and are only stable when they form a complex. A case in point is the interaction between the subunits of the hemoglobin molecule.
In contrast, non-obligate interactions are defined as interactions in which proteins are capable of existing independently but interact under specific conditions. Enzyme-inhibitor interactions serve as prototypical examples. Non-obligate interactions can be classified as either stable or transient based on their stability.
Stable Interactions vs. Transient Interactions
Stable PPIs result in the formation of long-lived complexes. Such interactions frequently serve as structural and functional components of cellular machinery. Remarkable examples include ribosomes and proteasomes.
In contrast, transient interactions are characterized by a relatively short lifespan, occurring and dissociating rapidly. Such interactions are frequently implicated in signal transduction and regulatory processes. Examples include kinase-substrate interactions and antigen-antibody interactions.
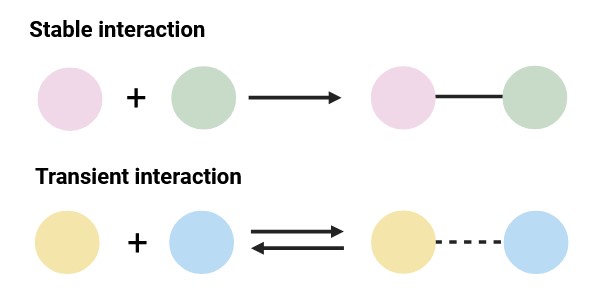 Figure 2. Classification of protein–protein interaction types based on stability (Created with BioRender.com).
Figure 2. Classification of protein–protein interaction types based on stability (Created with BioRender.com).
Covalent vs. Non-Covalent
Covalent interactions entail the transfer of electron pairs between atoms, thereby forming a robust and enduring bond. In the context of PPIs, covalent bonds are relatively uncommon but can be crucial for the stability and functionality of specific protein complexes. The principal categories of covalent interactions encompass disulfide bonds, isopeptide bonds, and cross-linking.
Non-covalent interactions are more prevalent and varied than their covalent counterparts. While they are typically less robust, these interactions are vital for the transient and reversible interactions that underpin numerous cellular processes. The principal categories of non-covalent interactions are hydrogen bonding, ionic (electrostatic) interactions, van der Waals forces, hydrophobic interactions, π-stacking, and cation-π interactions.
Examples of Crucial Protein-Protein Interactions
Ribosome Assembly: The ribosome is a complex molecular machine composed of ribosomal RNA and proteins. Its assembly involves numerous stable PPIs that are essential for protein synthesis.
Signal Transduction Pathways: Signaling pathways, such as the MAPK/ERK pathway, are dependent on transient interactions between kinases and their substrates to transmit signals from the cell surface to the nucleus. In this context, protein-ligand interactions represent a significant form of PPIs.
Immune Response: The interactions between antigens and antibodies are transient and highly specific, enabling the immune system to recognize and neutralize pathogens.
Cytoskeletal Dynamics: Actin and tubulin, two of the most important cytoskeletal proteins, interact with each other and with a variety of binding proteins in order to maintain cell shape, enable motility, and facilitate intracellular transport.
Analyzing and Predicting Protein-Protein Interactions
A number of techniques have been developed for the detection of protein interactions, which can also be employed for the prediction of PPIs. Furthermore, computational methods, including sequence-based, structure-based, machine learning, and AI-based approaches, have been shown to be highly effective for the prediction of PPIs.
| Techniques | Mechanism |
| Fluorescence Resonance Energy Transfer (FRET) | FRET measures the transfer of energy between two fluorophore-labeled proteins when they are in close proximity. |
| Surface Plasmon Resonance (SPR) | SPR measures changes in the refractive index near a sensor surface to which a protein is immobilized. The binding of another protein is detected in real time, providing kinetic data on the interaction. |
| Bio-layer Interferometry | This method measures the change in interference when a protein is added to a layer of another protein. |
| Nuclear Magnetic Resonance (NMR) Spectroscopy | NMR determines the spatial structure of a protein. |
| Yeast Two-Hybrid (Y2H) Assay | The Y2H assay detects PPIs by expressing potentially interacting proteins as fusions with the DNA binding and activation domains of a transcription factor in yeast. Interaction between the proteins reconstitutes the transcription factor and activates reporter gene expression. |
| Co-Immunoprecipitation (Co-IP) | Co-IP involves the use of an antibody to capture a protein of interest from a cell extract, along with its binding partners. The complexes are then analyzed by Western blotting or mass spectrometry. |
| Affinity Purification-Mass Spectrometry (AP-MS) | Tagged proteins are expressed in cells and purified along with their interacting partners. The complexes are then analyzed by mass spectrometry to identify the interacting proteins. |
| Bioinformatics Tools | Computational methods, including molecular docking and databases, predict and analyze PPIs based on known protein structures and interaction data. |
How Protein-Protein Interactions Drive Medical and Biotech Advances
Understanding PPIs is crucial for several reasons:
Synthetic Biology Tools: Engineering proteins with novel interactions has applications in synthetic biology, where designing new pathways or modifying existing ones can lead to the production of valuable biomolecules. For example, by designing biosensors with highly specific PPIs, it is possible to achieve high sensitivity and specificity in detecting target molecules, making them useful for medical diagnostics and environmental monitoring.
Drug Development: Targeting PPIs is a promising strategy in drug development. Small molecules or peptides that disrupt specific interactions can be used to modulate disease pathways, particularly in cancer and infectious diseases. For example, understanding the interaction between viral or bacterial proteins and host immune receptors helps to design effective vaccine candidates.
Disease Diagnosis and Prognosis: Aberrant PPIs are often associated with disease. Biomarkers based on such interactions may improve the diagnosis and prognosis of diseases like Alzheimer's and heart disease.
Cell-Based Therapy: In therapies like CAR-T cell therapy, engineered T cells rely on PPIs to recognize and kill cancer cells. The chimeric antigen receptor (CAR) on T cells must interact with antigens on cancer cells to initiate an immune response.
Protein-Based Therapy: Many therapeutic proteins function by interacting with other proteins. For example, monoclonal antibodies work by binding to specific antigens on the surface of cancer cells or pathogens, marking them for destruction by the immune system. PPIs are used to design therapeutic proteins with increased stability, specificity, and activity. Protein engineering can create variants that bind more effectively to their targets or evade immune detection.
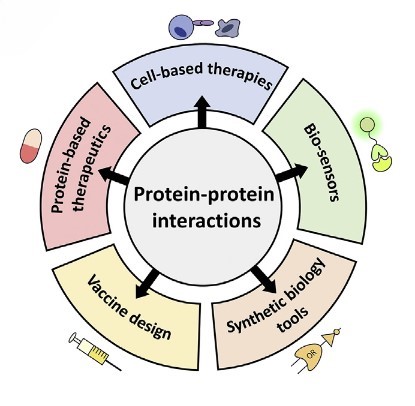 Figure 3. Overview of potential applications for novel PPIs (Marchand et al., 2022).
Figure 3. Overview of potential applications for novel PPIs (Marchand et al., 2022).
Creative Biostructure offers a comprehensive range of protein interaction services, leveraging our strong expertise, extensive experience, proprietary high-throughput screening and affinity ranking technology, and state-of-the-art equipment. Our analysis covers affinity ranking, measurement, and kinetics, providing valuable insights for constructing interaction maps, exploring sequence diversity, and examining posttranslational modifications, as well as facilitating rapid antibody screening. With high-precision data delivered at a reasonable cost and with speed, our services are tailored to meet your specific protein interaction analysis needs. For more information, please reach out to us.
References
- Acuner Ozbabacan, S. E., Engin, H. B., Gursoy, A., & Keskin, O. (2011). Transient protein-protein interactions. Protein Engineering Design and Selection, 24(9), 635–648.
- Marchand, A., Van Hall-Beauvais, A. K., & Correia, B. E. (2022). Computational design of novel protein–protein interactions – An overview on methodological approaches and applications. Current Opinion in Structural Biology, 74, 102370.
- Phizicky, E. M., & Fields, S. (1995). Protein-protein interactions: Methods for detection and analysis. Microbiological Reviews, 59(1), 94–123. https://doi.org/10.1128/mr.59.1.94-123.1995.