Protein Stability and Dynamics
Proteins, as fundamental biomolecules, play a critical role in virtually all biological processes. Their functionality is intrinsically linked to their structure, which must be maintained under various physiological conditions. Two key aspects that govern protein function are protein stability and protein dynamics.
Creative Biostructure provides a comprehensive suite of services to support researchers, pharmaceutical companies, and industrial partners in their quest to harness the full potential of proteins.
What is Protein Stability?
Protein stability refers to the ability of a protein to maintain its three-dimensional structure and functional state under varying environmental conditions. Stability includes both thermodynamic stability, which is the difference in free energy between the folded (native) and unfolded (denatured) states, and kinetic stability, which refers to the rate at which a protein unfolds or degrades. High stability means that a protein can resist denaturation and degradation over time.
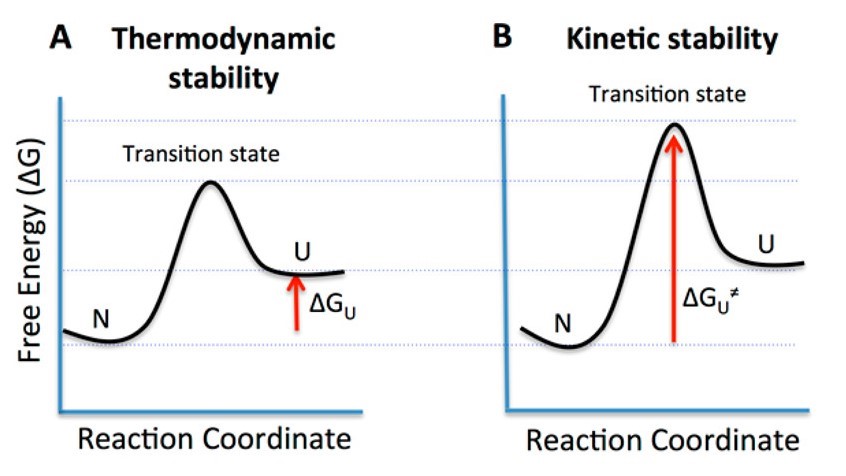 Figure 1. Free energy diagrams illustrating the different types of protein stability. (A) The thermodynamic stability of proteins is defined as the difference in free energy (∆GU) between the native (N) and unfolded (U) states. The low transition state free energy for unfolding causes the N and U states to exist in equilibrium, which favors the N state. (B) Kinetically stable proteins (KSPs) are characterized by having a high activation energy (∆GU≠) for unfolding that practically confines them in their native state (N). The stability of such proteins is under kinetic control, as their unfolding rate is very slow, which results in a longer half-life (Colón, et al., 2017).
Figure 1. Free energy diagrams illustrating the different types of protein stability. (A) The thermodynamic stability of proteins is defined as the difference in free energy (∆GU) between the native (N) and unfolded (U) states. The low transition state free energy for unfolding causes the N and U states to exist in equilibrium, which favors the N state. (B) Kinetically stable proteins (KSPs) are characterized by having a high activation energy (∆GU≠) for unfolding that practically confines them in their native state (N). The stability of such proteins is under kinetic control, as their unfolding rate is very slow, which results in a longer half-life (Colón, et al., 2017).
The Molecular Basis of Protein Stability
The stability of a protein is influenced by several molecular interactions and environmental factors:
Hydrophobic Interactions: The burial of hydrophobic residues in the protein core is a major driving force for protein folding and stability. These interactions help minimize the exposure of hydrophobic residues to the aqueous environment, stabilizing the folded state.
Hydrogen Bonds: Hydrogen bonds between backbone amides and carbonyls, as well as side chains, contribute to the secondary and tertiary structure of proteins. These interactions provide additional stabilization by linking different parts of the protein.
Electrostatic Interactions: Salt bridges and ionic interactions between charged residues stabilize the folded structure. The electrostatic environment of the solvent can also influence protein stability.
Van der Waals Forces: These weak interactions among closely packed atoms in the protein contribute to the overall stability by optimizing the packing of the protein interior.
Disulfide Bonds: Covalent disulfide bonds between cysteine residues can significantly increase protein stability, especially in extracellular proteins exposed to oxidizing environments.
Solvent Effects: The polarity and ionic strength of the solvent, as well as pH and temperature, can affect protein stability. Solvent molecules can participate in stabilizing or destabilizing interactions with the protein.
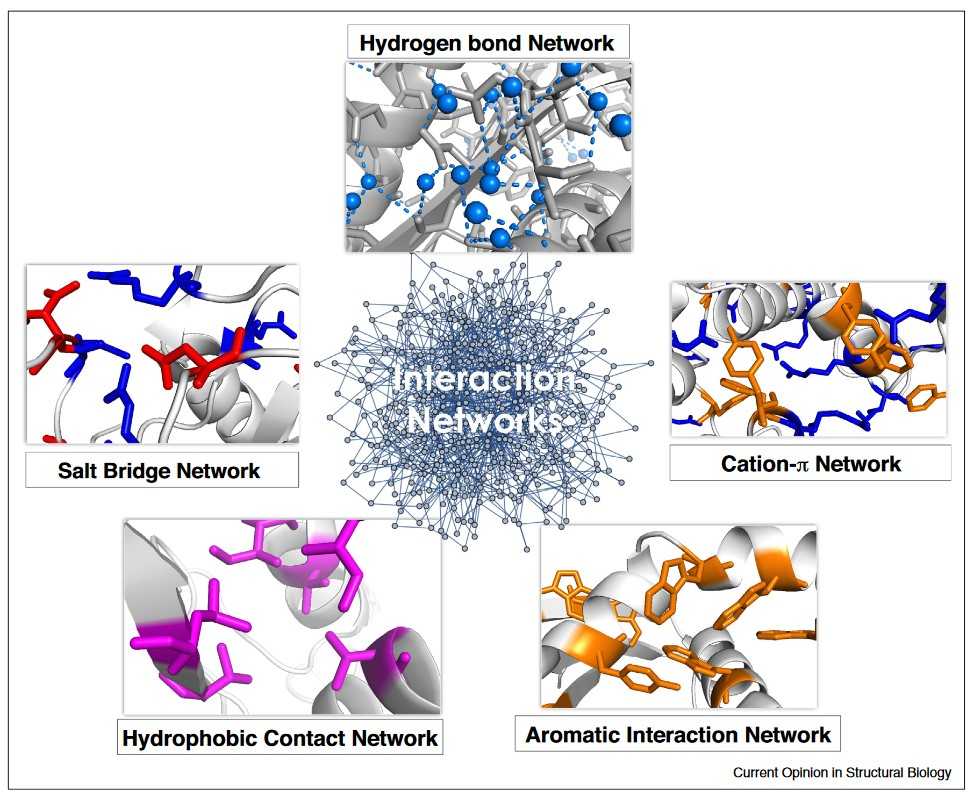 Figure 2. Schematic illustration of interaction networks that contribute to the stability of proteins (Pucci & Rooman, 2017).
Figure 2. Schematic illustration of interaction networks that contribute to the stability of proteins (Pucci & Rooman, 2017).
What is Protein Dynamics?
Protein dynamics refers to the motions within a protein molecule that contribute to its function. These motions can range from small atomic fluctuations to larger conformational changes and domain motions. Dynamics are essential for several biological activities, including enzyme catalysis, signal transduction, and molecular recognition. Protein dynamics occur on a variety of timescales, from femtoseconds to seconds or longer, reflecting the complex nature of protein motions.
A Hierarchical Strategy for Protein Dynamics
The hierarchical strategy of protein dynamics refers to the organization of protein motions into different levels, each characterized by different timescales and amplitudes. This hierarchical approach recognizes that protein dynamics are not uniform but occur over a wide range of spatial and temporal scales, from rapid local fluctuations to slower, large-scale conformational changes. Understanding these hierarchical levels is critical to understanding how proteins function in a dynamic cellular environment. Here's a closer look at the hierarchical strategy of protein dynamics:
Atomic Fluctuations
Atomic fluctuations are the fastest and smallest motions within proteins, on the scale of femtoseconds to picoseconds. They involve vibrations and rotations of individual atoms or small groups of atoms about their equilibrium positions. These motions contribute to the fine-tuning of protein structures, influence hydrogen bonding networks, and affect interactions with ligands and other molecules.
Side Chain Motions
Side chain motions involve the movement of amino acid side chains on a timescale of picoseconds to nanoseconds. These can be rotations around single bonds (such as the chi angles in amino acids) or more substantial rearrangements of side chains. Side chain motions are critical for the flexibility of active sites in enzymes, the binding of substrates and inhibitors, and the formation and disruption of transient interactions with other proteins or molecules.
Loop and Secondary Structure Motions
These motions involve larger segments of the protein, such as loops, α-helices, and β-sheets. They include bending, twisting, and fluctuations of these structural elements. These motions last from nanoseconds to microseconds. Loop and secondary structure motions are essential for processes such as ligand binding and release, allosteric regulation, and the adaptation of protein structures to their functional states.
Domain Motions
Domains are relatively independent folding units within proteins. Domain motions involve the movement of these large segments relative to each other and last from microseconds to milliseconds. They can include hinge-like motions, shear motions, and rotational adjustments. Domain motions play a critical role in the function of multi-domain proteins such as kinases, motor proteins, and receptors. These motions facilitate processes such as signal transduction, molecular recognition, and enzyme catalysis.
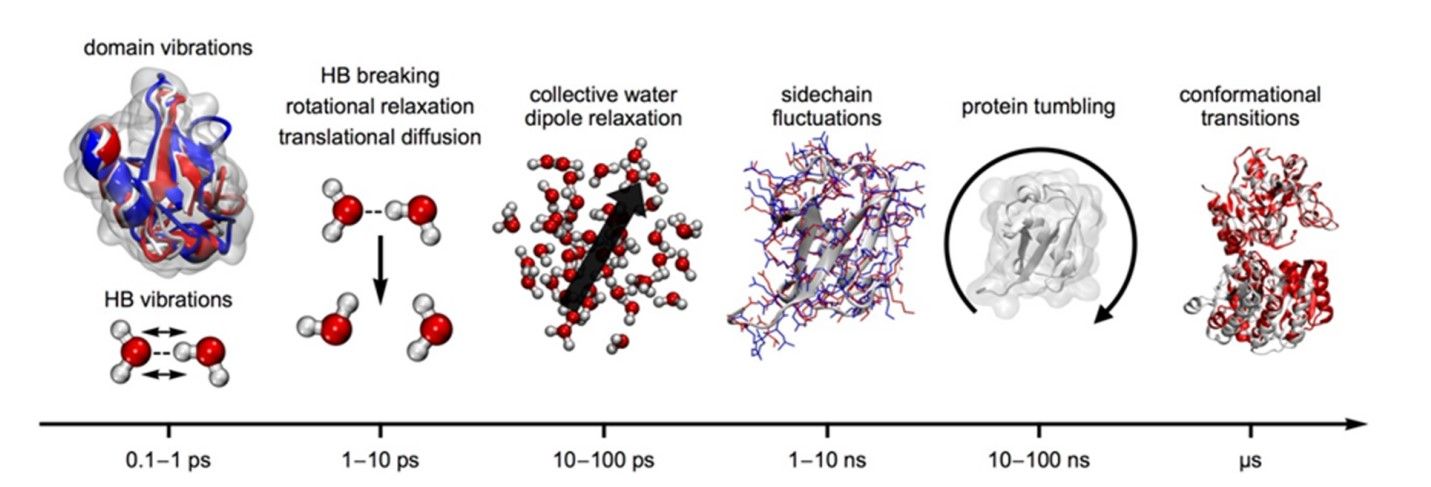 Figure 3. Hierarchy of time scales for protein motions. From left to right: (0.1–1.0 ps/1–10 THz) intramolecular vibrations of protein domains and secondary structure elements, including skeleton motions and intermolecular vibrations of the water hydrogen bond network; (1–10 ps/0.1–1 THz) relaxation processes in the hydrogen bond network of water, i.e., hydrogen bond rearrangements, single molecule rotation, and translational diffusion; (10–100 ps/10–100 GHz) collective dipole relaxation in water; (1–10 ns/0.1–1 GHz) protein side chain fluctuations; (10–100 ns/10–100 MHz) protein rotational tumbling motions; (µs/MHz) conformational transitions in proteins, shown as a hinge motion of glycogen synthase (Xu & Havenith, 2015).
Figure 3. Hierarchy of time scales for protein motions. From left to right: (0.1–1.0 ps/1–10 THz) intramolecular vibrations of protein domains and secondary structure elements, including skeleton motions and intermolecular vibrations of the water hydrogen bond network; (1–10 ps/0.1–1 THz) relaxation processes in the hydrogen bond network of water, i.e., hydrogen bond rearrangements, single molecule rotation, and translational diffusion; (10–100 ps/10–100 GHz) collective dipole relaxation in water; (1–10 ns/0.1–1 GHz) protein side chain fluctuations; (10–100 ns/10–100 MHz) protein rotational tumbling motions; (µs/MHz) conformational transitions in proteins, shown as a hinge motion of glycogen synthase (Xu & Havenith, 2015).
The Roles of Protein Stability and Dynamics
The stability and dynamics of proteins are fundamental to their physiological functions:
Enzyme Function: Enzymes rely on a stable active site to catalyze reactions efficiently. However, dynamic motions within the enzyme are critical for substrate binding, product release, and allosteric regulation.
Signal Transduction: Protein dynamics allow for conformational changes necessary for signal transduction across membranes or within the cell. For example, receptor proteins often undergo dynamic rearrangements upon ligand binding.
Protein-Protein Interactions: Stable yet flexible binding interfaces are essential for transient interactions between proteins. These interactions underlie many cellular processes, including metabolic pathways and cellular assembly processes.
Molecular Recognition: Antibodies and other recognition proteins must be stable to effectively bind their targets. However, dynamic flexibility is often required for high-affinity binding and specificity.
Protein Degradation and Turnover: The regulated degradation of proteins is critical for maintaining cellular homeostasis. Proteins of lower stability are more susceptible to degradation, which is often mediated by proteasomes and other degradation pathways.
The Relationship Between Protein Stability and Dynamics
Protein stability and dynamics are intrinsically linked and influence each other. A stable protein must retain some degree of flexibility to perform its functions, while dynamic proteins must maintain sufficient stability to avoid denaturation. This balance is critical for the proper functioning of proteins within the cellular environment.
For example, enzymes must have a stable active site for catalysis, but also need dynamic movements for substrate binding and product release. Similarly, allosteric proteins require stable conformational states, but must also undergo dynamic transitions to respond to effector molecules.
Assay Methods for Protein Stability and Dynamics
Several techniques are used to study protein stability and dynamics:
| Techniques | Attribute measured | Characteristics Indicated |
| Differential Scanning Calorimetry (DSC) | Heat capacity of a protein | Thermal stability and unfolding transitions |
| Circular Dichroism (CD) Spectroscopy | The secondary structure of proteins Changes in response to temperature, pH, or other conditions |
Protein stability |
| Molecular Dynamics (MD) Simulations | Computational simulations: Atomic-level motions and stability of proteins over time | Protein stability |
| Atomic force microscopy (AFM) | Detailed information on the dynamics and mechanical stability of individual protein molecules | Protein dynamics and mechanical stability |
| Nuclear Magnetic Resonance (NMR) Spectroscopy | Relaxation times, chemical shifts, and nuclear Overhauser effects (NOEs) | Protein dynamics |
| X-ray Crystallography | β-factors and multiple conformations within the crystal | Protein dynamic regions |
At Creative Biostructure, we focus on delivering in-depth protein stability and dynamics assays, crucial for various fields including science, medicine, and industry. Our expertise uncovers the inner workings of proteins, revealing how they interact and adapt to different conditions. This knowledge is key for drug development, protein design, disease research, structural biology, and industrial innovation. If you're interested in how our services can enhance your protein stability and dynamics research, don't hesitate to contact us for a personalized quote.
Reference
- Colón, Wilfredo, et al. Biological Roles of Protein Kinetic Stability. Biochemistry, vol. 56, no. 47, Nov. 2017, pp. 6179–86.
- Pucci, Fabrizio, and Marianne Rooman. Physical and Molecular Bases of Protein Thermal Stability and Cold Adaptation. Current Opinion in Structural Biology, vol. 42, Feb. 2017, pp. 117–28. PubMed.
- Xu, Y., & Havenith, M. (2015). Perspective: Watching low-frequency vibrations of water in biomolecular recognition by THz spectroscopy. The Journal of Chemical Physics, 143(17), 170901.
