Structural Research of Lipopolysaccharide (LPS) Transport Proteins
Lipopolysaccharides (LPS) play a crucial role in the outer membrane of Gram-negative bacteria, acting as a robust physical barrier against environmental threats and antibiotics. Understanding the structure and mechanism of LPS transport proteins, particularly the LptD/E protein complex involved in LPS insertion, is of utmost importance in advancing our knowledge of bacterial defense mechanisms and developing effective antibacterial strategies. In recent years, significant progress has been made in the structural research of LPS transport proteins, thanks to advancements in cryo-electron microscopy (cryo-EM) and X-ray crystallography techniques.
Cryo-EM Structural Insights
A recent study presents a detailed cryo-EM structure of a partially opened LptDE transporter complex at an impressive resolution of 3.4 Å. This breakthrough allowed us to visualize the complex in unprecedented detail, providing insights into the transitional state during LPS release.
Moreover, a subset of particles in this study enabled the modeling of a laterally fully opened LptDE complex. These findings provide novel information about the dynamic nature of LptD transporter and its significance in LPS insertion and bacterial outer membrane biogenesis.
Rigid Chaperones Derived from Nanobodies Enhancing Structural Determination
The success of this cryo-EM study was further bolstered by the incorporation of rigid chaperones derived from nanobodies. These nanobody-based chaperones acted as stabilizing agents, preserving the partially opened LptDE complex during the imaging process. The result was an improved structural determination, allowing us to gain deeper insights into the conformational changes of LptDE during the LPS transport process.
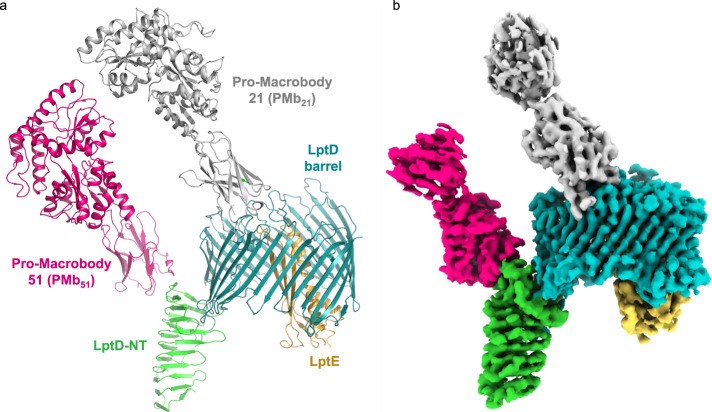 Figure 1. Cryo-EM structure of the NgLptDE-PMb21/PMb51 complex. (Botte M, et al., 2022)
Figure 1. Cryo-EM structure of the NgLptDE-PMb21/PMb51 complex. (Botte M, et al., 2022)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| LptD-LptE lipopolysaccharide transport complex (expressed in Escherichia coli) | Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 | X-ray diffraction | 2.80 Å | 4N4R |
| LptD-LptE lipopolysaccharide transport complex (expressed in Escherichia coli) | Shigella flexneri | X-ray diffraction | 2.39 Å | 4Q35 |
| LptE lipopolysaccharide transport protein (expressed in Escherichia coli) | Escherichia coli K-12 | X-ray diffraction | 2.34 Å | 4NHR |
| PgaA exopolysaccharide transporter (expressed in Escherichia coli) | Escherichia coli K-12 | X-ray diffraction | 2.82 Å | 4Y25 |
| LptD-LptE lipopolysaccharide transport complex (expressed in Escherichia coli) | Yersinia pestis | X-ray diffraction | 2.75 Å | 5IXM |
| LptD-LptE lipopolysaccharide transport complex (expressed in Escherichia coli) | Klebsiella pneumoniae | X-ray diffraction | 2.94 Å | 5IV8 |
| LptD-LptE lipopolysaccharide transport complex (expressed in Escherichia coli) | Pseudomonas aeruginosa PAO1, Pseudomonas aeruginosa | X-ray diffraction | 2.99 Å | 5IVA |
| LptD-LptE lipopolysaccharide transport complex, in complex with ProMacrobodies (expressed in Escherichia coli) | Neisseria gonorrhoeae, synthetic construct, Methanosarcina mazei | Cryo-EM single particle analysis | 3.40 Å | 7OMM |
Table 1. Structural research of LPS transport proteins.
With years of experience in the biotechnology industry, Creative Biostructure stands as a renowned and reliable partner for structural analysis services. Our group of skilled scientists employs state-of-the-art techniques, including cryo-electron microscopy (cryo-EM), X-ray crystallography, and NMR spectroscopy, to acquire structural data with high resolution. Contact us to explore how our advanced capabilities can enhance your research endeavors and propel you toward accomplishing your scientific objectives.
References
- Wang Y, et al. Structural basis for translocation of a biofilm-supporting exopolysaccharide across the bacterial outer membrane. Journal of Biological Chemistry. 2016, 291(19): 10046-10057.
- Botte M, et al. Cryo-EM structures of a LptDE transporter in complex with Pro-macrobodies offer insight into lipopolysaccharide translocation. Nature Communications. 2022, 13(1): 1826.