Epitope Mapping
At present, cryogenic electron microscopy (cryo-EM) combined with 3D reconstruction technology has been able to simulate the structure of assembled macromolecules. It can also be used for epitope mapping in order to localize proteins or detect immunogenicity, making structural biology into a new era. Epitopes can be divided into linear epitopes and conformational epitopes. Their differences are as follows:
- Linear epitopes are composed of linear peptide chains of about 5 to 20 amino acids in proteins, while conformational epitopes are composed of amino acids that are not continuous in the protein sequence and come together during the 3D folding of the protein.
- Linear epitopes are also referred to as continuous epitopes because the antibody binding site and its amino acid side chain skeleton form a strong diversity of structure and are linked to adjacent peptide chains. General protein epitope mapping is also the easiest method to find linear epitope; The conformation epitopes are difficult to locate and are suitable for mapping large protein regions.
Currently, protein epitope mapping plays an important role in the structural analysis of antigen-antibody complexes, such as SARS-CoV-2. For example, human neutralizing antibodies (hNAbs) targeting the SARS-CoV-2 spike protein in the host ACE2 receptor-binding domain (RBD) have been found in many laboratories to show therapeutic promise. By mapping the proteins of the complex formed by different neutralizing antibodies with SARS-CoV-2, the researchers provided insight into the likelihood that SARS-CoV-2 could escape from antibodies that are induced during infection or delivered therapeutically. It provides various useful insights for the current and future classification of targeted antibodies and possible immune responses to SARS-CoV-2.
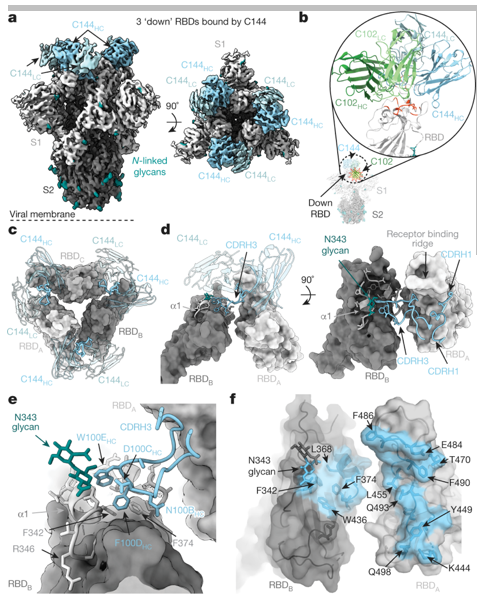 Figure 1. Cryo-EM structure of the C144-S complex illustrates a distinct VH3-53 NAb binding mode. (Barnes C O, et al. 2020)
Figure 1. Cryo-EM structure of the C144-S complex illustrates a distinct VH3-53 NAb binding mode. (Barnes C O, et al. 2020)
We are committed to developing cryo-EM solutions for protein epitope localization. The general workflow is shown as below:
| Cryo-EM Sample Preparation | The EM girds are prepared by plasma treatment, sample deposition and vitrification using a repeatable biological sample glass table to ensure the structural integrity of the vitrification sample and no contamination. |
| Imaging and Data Collection | The prepared grids are imitated on a TEM. After data collection, the images are used to generate cryo-EM maps of the proteins. |
| Data Sorting | The cryo-EM maps are processed to find a suitable template, and the code is used to search the protein library, so as to identify the residues on the binding interface and compare the similarity. And we can combine the crystal structures to reconstruct the model. |
| Model Fitting | By moving or rotating the protein structure, the model is fitted to the cryo-EM diagram, and epitope mapping can be achieved. |
If you are interested in our epitope mapping service, please feel free to contact us. We are looking forward to cooperating with you.
Ordering Process
Reference
- Barnes C O, Jette C A, Abernathy M E, et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020, 588(7839): 682-687.

