Epitope Mapping Services
Creative Biostructure provides various techniques for mapping of conformational epitopes using the most advanced structural biology technology at high resolution. Technologies have been developed rapidly used for studying the interactions of antibodies with specific regions of protein antigens, especially for conformational epitopes. Important applications of epitope mapping are found within the area of immunochemistry.
Determining the precise epitope of therapeutic antibodies is beneficial in understanding the structure–activity relationship of the drug, but in many cases is not done due to the structural complexity of, in particular, conformational protein epitopes. The screening of phage libraries displaying random peptides has had limited impact on the mapping of conformational epitopes because the selected peptides are regarded as mimotopes of the genuine epitope of the antibody's interacting antigen. X-ray co-crystal structure of the antigen–antibody complex remains the gold standard of epitope determination, other widely available technologies include cryo-EM and NMR.
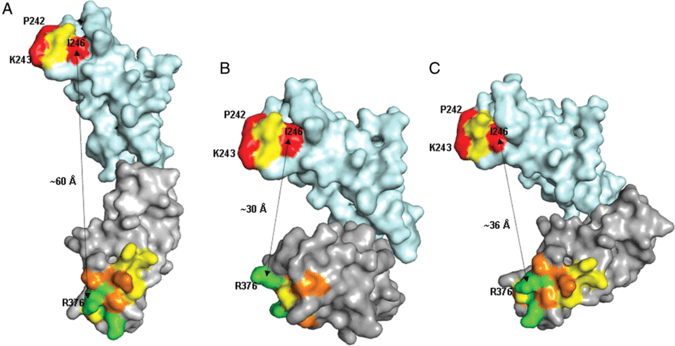 Figure 1. Alternative arrangements of C2 (cyan) and C3 (grey) domains of CD22 obtained by superposition of the models.
Figure 1. Alternative arrangements of C2 (cyan) and C3 (grey) domains of CD22 obtained by superposition of the models.
Technologies for conformational epitope mapping provided by Creative Biostructure:
1. Co-crystallization is always feasible if obtaining high concentrations of good-quality proteins after finding the correct conditions to produce diffracting crystals. This approach is technically challenging, requires large amounts of purified protein, and can be time-consuming and expensive.
2. NMR epitope mapping can determine structures and epitopes of proteins, protein-ligand complexes, peptides, macrocycles, etc. This method provides more detailed information than mutagenesis or peptide mapping and can be much more rapid than X-ray crystallography.
3. cryo-EM epitope mapping service is provided based on our efficient and robust platform to perform epitope mapping of antibody- antigen interactions using high-throughput Electron Microscopy (EM) and single particle analysis. This method is applicable to proteins in the range of 100kDa to 4MDa as well as both symmetric and asymmetric complexes at high resolution.
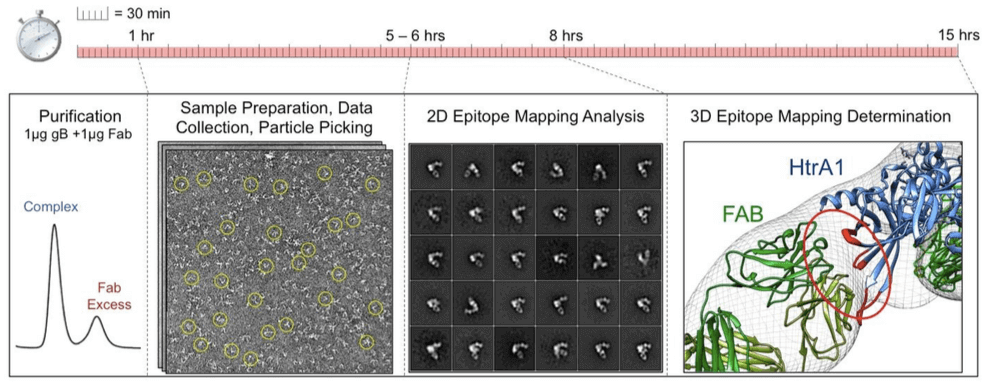 Figure 2. Schematic representation of the EM data collection and processing pipeline.
Figure 2. Schematic representation of the EM data collection and processing pipeline.
Our technical support team has significant experience in diverse biological fields (structural biology, membrane protein, protein engineering, virology, etc.). Please contact our experts with your project of interest. We look forward to discussing your ideas, answering questions, and helping you design a tailored proposal for your epitope discovery needs at high resolution.
Ordering Process
References:
- D. Bannister, et al. Epitope mapping and key amino acid identification of anti-CD22 immunotoxin CAT-8015 using hybrid β-lactamase display. Protein Eng Des Sel. 2011 Apr; 24(4): 351–360.
- Alberto Estevez, et al. EM by EM: High-Efficiency Epitope Mapping using High-Throughput Electron Microscopy. Microsc. Microanal. 22 (Suppl 3), 2016.
