Truncation Libraries
With abundant experience in gene libraries construction, Creative Biostructure has established an advanced mutagenesis library construction platform. We provide high-quality truncation libraries construction services to meet all of customers’ requirements.
A truncation library is produced by a systemic truncation of the peptide's sequence from each terminus and it can help researchers to determine the minimum length demanded for protein activity. Truncation libraries enable to decide the role of the sequences surrounding key residues, this goes after the identification of key residues by alanine scanning library studies. On various occasions, the proteolytic stability of peptides can be increased by truncation library screening. Truncation library has become a powerful tool to study the extent to which peptide drugs undergo metabolic degradation, which is a vital consideration in bringing drugs to the market.
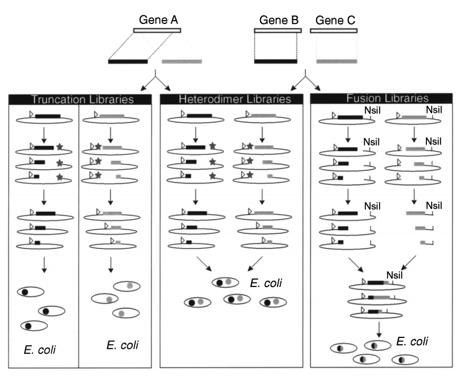 Figure 1. Combining incremental truncation libraries.
Figure 1. Combining incremental truncation libraries.
- Approaches for Truncation Libraries Construction
Incremental truncation is a common technology to generate truncation library, which can produce a combinatorial library including every onebase-pair deletion of a gene or gene fragment and does so in the lack of detailed structural information. With the development of this technology, Creative Biostructure has developed several approaches to create incremental truncation libraries: via the combination of α-phosphothioate dNTPs by PCR, through a time-dependent nuclease approach and by primer extension (THIO (pcr) truncation and THIO (extension) truncation, severally). Truncation peptide libraries are served as an essential part in authenticating the shortest amino acid sequence required for activity.
- Applications of Truncation Libraries
The incremental truncation technology has a plenty variety of applications in protein engineering. Firstly, it enables to create a library of carboxy-terminal or amino-terminal truncations of a simplex gene. By this means, connections between sequence, structure and function are effectively defined by such protein fragments, involving the detection of regions responsible for protein–protein interactions. In addition, the stochastic pairing of members of such libraries is used to confirm protein fragments which can non-covalently combine to form functional proteins. This approach for protein fragment complementation can be regarded as a tactics for the transformation of a monomeric protein into a heterodimer. Eventually, integrated fusion libraries between fragments of two genes can be produced by incremental truncation. This means has been applied in protein engineering, especially in the production of assorted fusion libraries between genes which have low levels of homology.
Creative Biostructure provides various protein engineering and analysis services, including protein evolution. Please feel free to contact us to discuss your project!
Ordering Process
References:
Lutz S, Ostermeier M, Benkovic S J. Rapid generation of incremental truncation libraries for protein engineering using α-phosphothioate nucleotides[J]. Nucleic Acids Research, 2001, 29(4): e16-e16.
Ostermeier M, Nixon A E, Shim J H, et al. Combinatorial protein engineering by incremental truncation[J]. Proceedings of the National Academy of Sciences, 1999, 96(7): 3562-3567.
Qian Z, Horton J R, Cheng X, et al. Structural redesign of lipase B from Candida antarctica by circular permutation and incremental truncation[J]. Journal of molecular biology, 2009, 393(1): 191-201.
