Super-resolution Localization Microscopy Service
Optical microscopes have been used to observe the microscopic biological world. However, the traditional optical microscope is limited by resolution and cannot observe smaller biomolecules and structures. With the emergence of scanning electron microscopy, scanning tunneling electron microscopy, and atomic force microscopy, it has become possible to achieve nanoscale resolution. Still, these techniques are destructive to samples and can only observe the surface, which is unsuitable for biological samples. To solve this problem, Creative Biostructure provides a variety of super-resolution imaging techniques suitable for imaging biological samples, including ground state depletion followed by individual molecule return (GSDIM), photoactivated localization microscopy (PALM), stochastic optical reconstruction microscopy (STORM), etc., to help understand biological structures and systems, and assist customers' research.
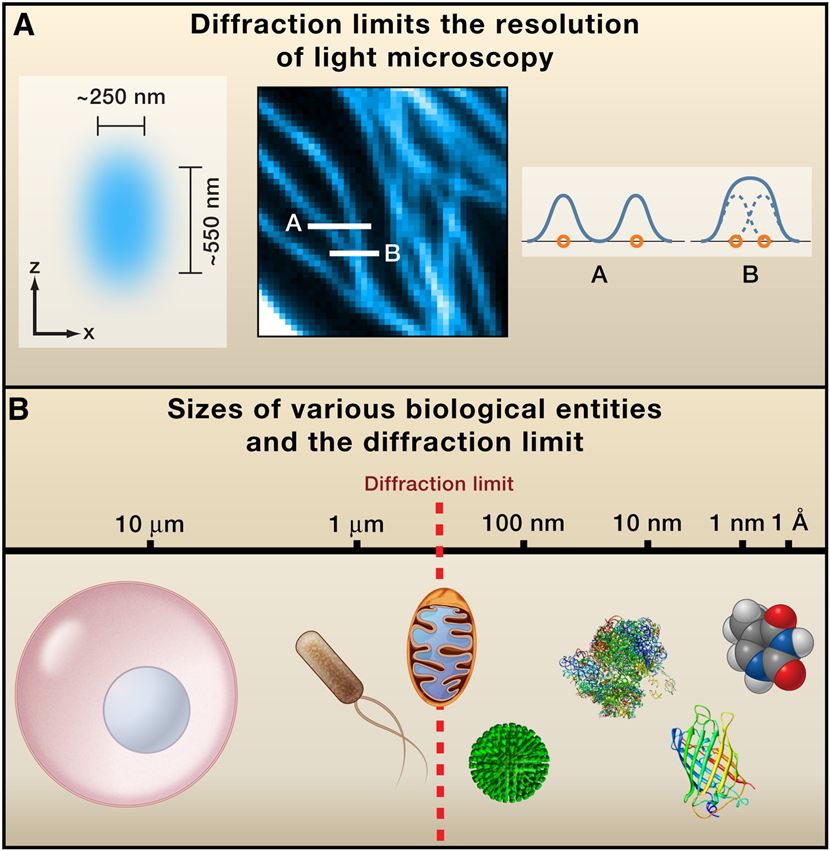 Figure 1. Diffraction-Limited Resolution of Conventional Light Microscopy.
Figure 1. Diffraction-Limited Resolution of Conventional Light Microscopy.
Our Super-resolution Localization Microscopy Techniques
Ground State Depletion Followed by Individual Molecule Return (GSDIM)
The key to overcoming the 200 nm diffraction limit with GSDIM microscopy is sequentially turning on and off and temporally separating fluorophores. The electrons of fluorophores are accumulated in a metastable, non-fluorescent dark state (triplet state and others) by a high-power laser beam. Molecules return stochastically to the ground state from the dark state for re-entering the fluorescence cycles called "blinking.” These fluorescent blinking events are captured over time. The diffraction-limited events are read out by applying a fitting algorithm to get a high-resolution image. Eventually, the ultimate high-resolution image is reconstructed by plotting the measured positions of thousands of recorded events.
Photoactivated Localization Microscopy (PALM)
The principle of PALM is to "switch" the light-activated fluorescent protein combined with the target molecule, locate it in batches, and determine the position of the central light spot. The probability of fluorescent proteins being activated is proportional to the intensity of the excitation light; as long as the activated proteins are few enough and far enough apart, they can be localized. After data acquisition, these fluorescent proteins are photobleached until they are completely inactivated, and then other fluorescent proteins are activated for localization. By repeating this process, all target molecules in the sample can be localized, and these raw data can be combined to obtain a super-resolution image of the target molecule.
Stochastic Optical Reconstruction Microscopy (STORM)
The basic principle of STROM is like that of PALM, except that instead of photoactivatable fluorescent proteins, STORM uses synthetic photoconvertible fluorescent dyes to bind to target molecules. Fluorescent dyes are excited into an emission state and then enter a dark state, where they combine with free oxygen and enter a bleached state. In the bleached state, the dye does not fluoresce again. If the dye is not allowed to combine with free oxygen, it cannot enter the bleached state and remain dark. High-power excitation light can make the dye re-enter the emissive state from the dark state. This switching from light to dark and back again looks like the dye is "flickering.” STORM performs super-resolution positioning by randomly "lighting" target molecules in batches and collects data faster.
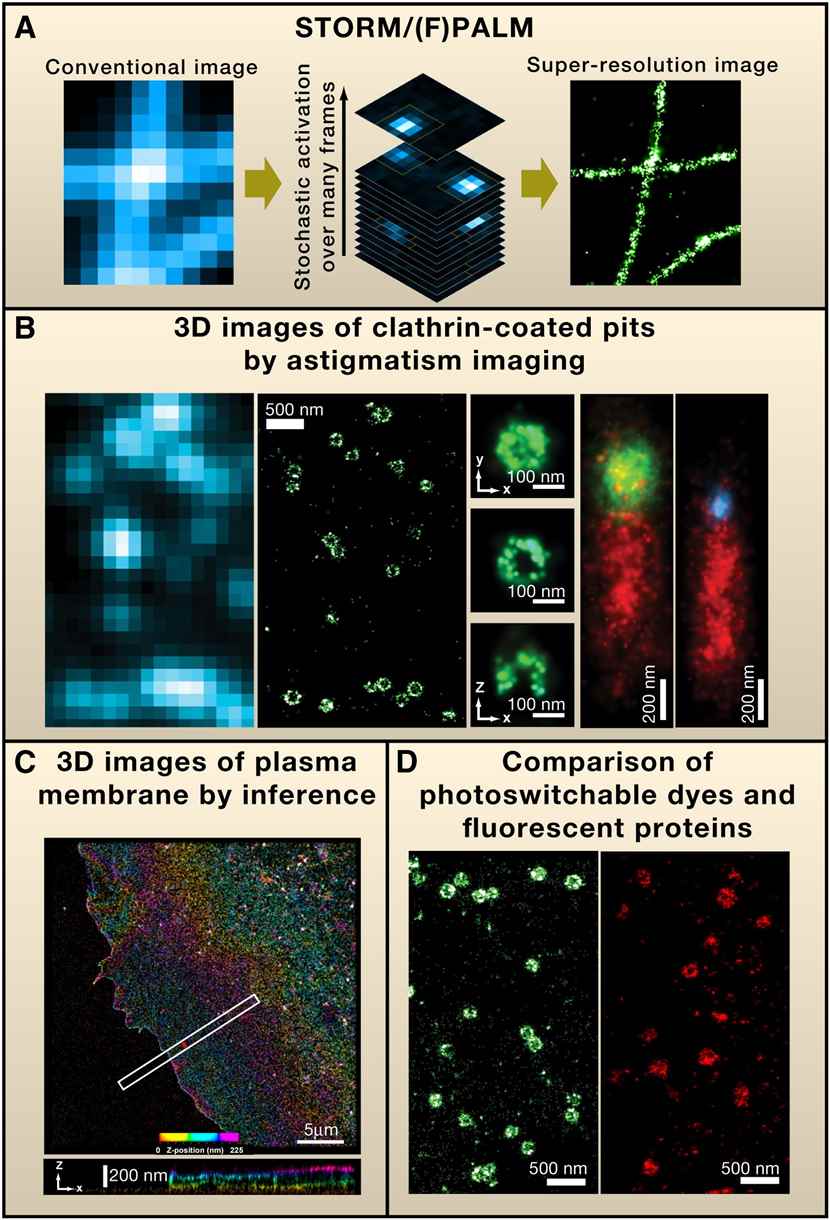 Figure 2. Super-resolution Fluorescence Microscopy by Single-Molecule Switching.
Figure 2. Super-resolution Fluorescence Microscopy by Single-Molecule Switching.
Applications of Super-resolution Localization Microscopy Service
- Isolation of exosomes from different sources
- Exosome Imaging and Tracking
- Observation of ultrastructure of various dyed, non-stained, and fluorescently labeled tissues and cells
- Observation of protein localization and co-localization in multicolor fixed and living specimens
- Observation of dynamic changes in the ultrastructure of living cells
- 3D imaging of fixed specimens
- Time series collection of living specimens
- Observation of cell membrane surface signals
- Single-molecule imaging
Creative Biostructure promises to work closely with customers, please feel free to contact us for more information or a detailed quote.
Ordering Process
References
- Huang B., et al. Breaking the diffraction barrier: super-resolution imaging of cells. Cell. 2010, 143(7): 1047-1058.
- Ovesný M., et al. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics. 2014, 30(16): 2389-2390.
- Rust M J., et al. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nature Methods. 2006, 3(10): 793-796.
- Ma H, Liu Y. Super-resolution localization microscopy: Toward high throughput, high quality, and low cost. APL Photonics. 2020, 5(6).
