Membrane Protein Structure Determination by Solid-state NMR
The biomolecule structures are most commonly achieved by X-ray crystallography, solution-state Nuclear Magnetic Resonance (NMR) spectroscopy, and cryo-electron microscopy (cryo-EM). To reveal the bioprocess mechanism, it is important to determine the native structure and intact assemblies of biomolecules in the physiological environment. X-ray crystallography is one of the most well-established techniques for structural biology; and more than 130 thousand biomolecule structures have been solved by X-ray crystallography. However the biggest limitation is that it requires the specimen to be properly crystalized and crystal grow in largely and orderly. Along with the fast development of direct electron detectors and image processing algorithms, cryo-EM has emerged to be a powerful tool for 3D structure characterization, with increased resolution and minimal sample preparation. With Cryo-EM, high-resolution structures can be obtained for molecular weights ranging from 120 kDa-11 MDa. However, the resolution of cryo-EM is limited for biomolecules with conformational heterogeneity and flexibility.
Solid-state NMR (ssNMR) provides rich information on the structure, dynamics, and interactions of molecules and complexes; and therefore has more advantages in resolving small protein structures and obtaining dynamic configurations. Moreover, due to the development of cross polarization (CP) and magic angle spinning (MAS) methodology, ssNMR can be applied to molecules with a wider range of molecular weight, allowing reconstruction of atomic-resolution structures.
Many membrane proteins, especially transporters and receptors, are targets of antibodies. Therefore, the structural characterization of intact assemblies, and revealing of the inter-molecular interactions is essential in the process of drug discovery. It has been reported that ssNMR provides structural details of membrane proteins embedded within lipids. Investigation of drug candidates and their functional pathways will be highly accelerated by a combination ssNMR data with solution-state NMR, X-ray crystallography, scanning transmission electron microscopy, fiber diffraction, and small-angle scattering techniques.
Creative Biostructure provides different types of ssNMR rotors for increasing the sensitivity of signal and reaching of extremely high spinning frequencies. Samples are operated under magic-angle spinning (MAS) conditions with common procedures for getting high resolution NMR spectra data (Figure 1).
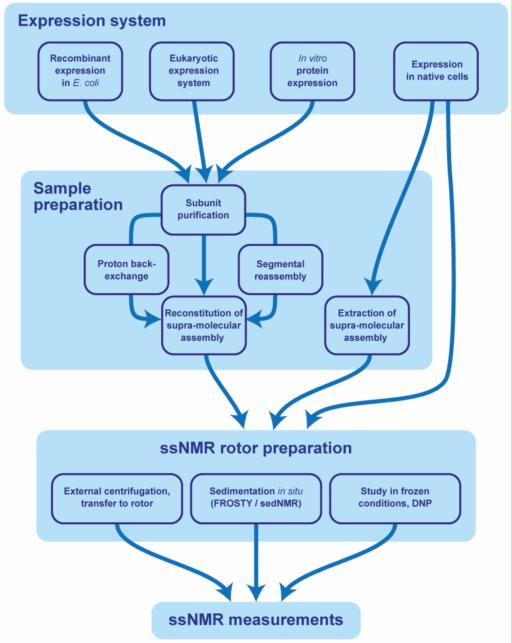 Fig. 1. Sample preparation for the ssNMR study of supra-molecular assemblies.
Fig. 1. Sample preparation for the ssNMR study of supra-molecular assemblies.
Creative Biostructure promises to work closely with our customers to provide excellent services from protein preparation to ssNMR data collection and analysis. We are dedicated to helping our customers achieving their goal.
Please feel free to contact us for a detailed quote.
Ordering Process
References:
- Jean-Philippe Demers, Pascal Fricke et. al. Structure Determination of Supra-Molecular Assemblies by Solid-State NMR: Practical Considerations. Prog Nucl Magn Reson Spectrosc, 2018;109:51-78.
- Anthony Watts, Ian J. Burnett, et.al. Membrane protein structure determination by solid state NMR. Nat. Prod. Rep., 1999; 16:419–423.
