Model Reconstruction of Crystal Structures Using Cryo-EM and NMR
Cryo-Electron Microscopy (cryo-EM), Nuclear Magnetic Resonance (NMR) spectroscopy and X-ray crystallography are routinely used to determine structures of macromolecules. Due to the fact that crystallization is not required in structural analysis and only a small amount of sample is needed, cryo-EM has quickly gained popularity in structural biology. With the advance of microscope design, imaging hardware and image processing, cryo-EM has become one of the most powerful tools to solve the structure of biomolecules. However, it is a major bottleneck that obtaining structures at very high resolution can be something challenging as to support the development of new drugs. Therefore, combination of cryo-EM with X-ray crystallization or NMR has become the complementary approaches to structural biology and drug discovery.
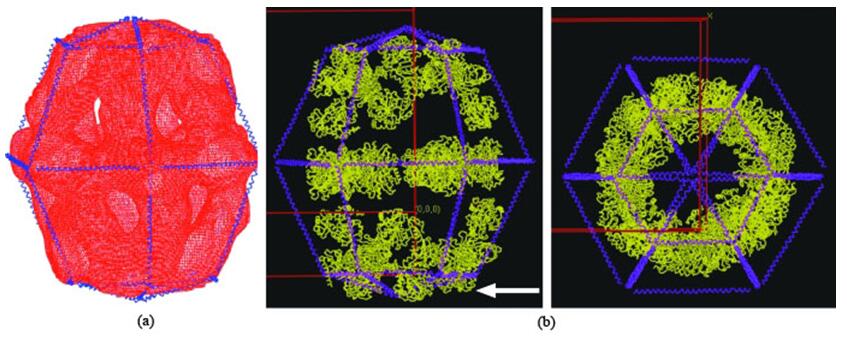 Figure 1. Structure of yeast fatty-acid synthase (FAS).
Figure 1. Structure of yeast fatty-acid synthase (FAS).
In this approach, a low-resolution structure is determined by three-dimensional (3D) Cryo-EM, which is used as the initial searching model for x-ray crystallization or NMR (Figure 1). After phasing the high-resolution X-ray data with low-resolution EM reconstruction by molecular replacement, theoretical phases can be calculated. This approach has been proved successful in determining the structures of some large complexes such as viruses, ribosomes, and others (Figure 2 and Figure 3).
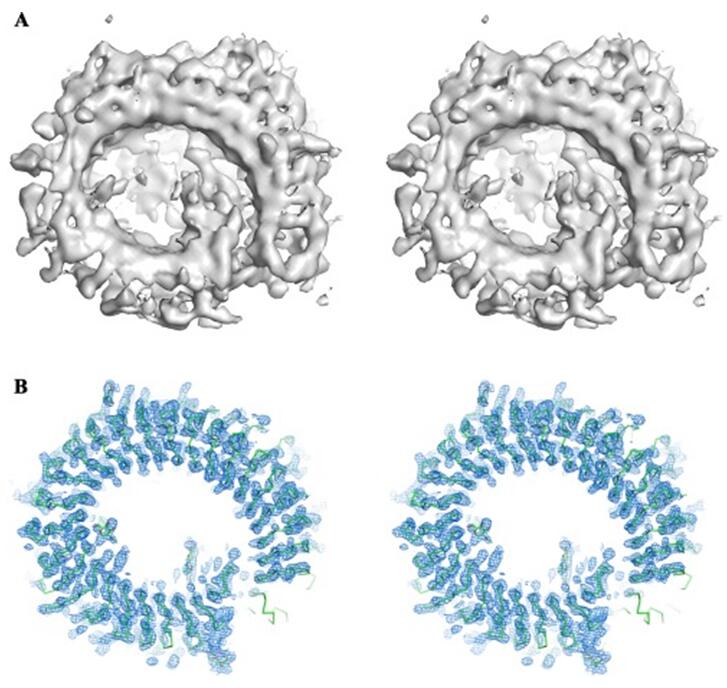 Figure 2. Cryo-EM 3D reconstruction helps phasing in X-ray crystallography.
Figure 2. Cryo-EM 3D reconstruction helps phasing in X-ray crystallography.
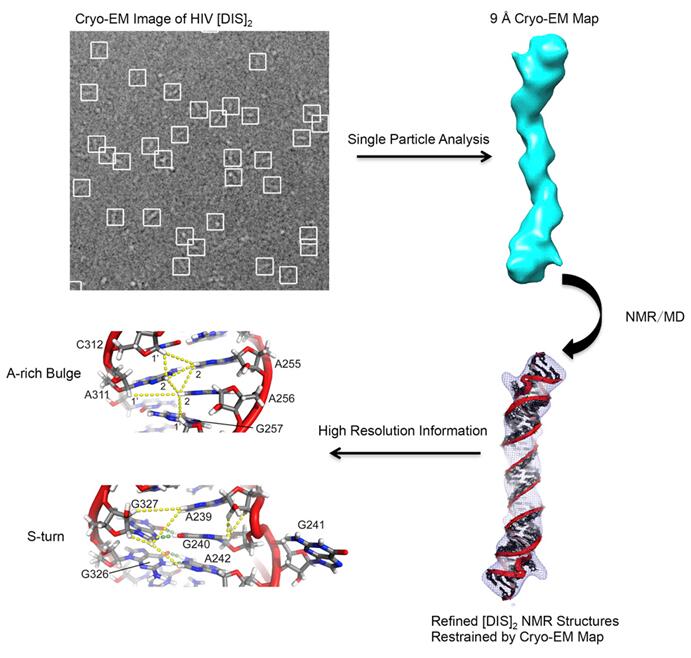 Figure 3. Atomic model of HIV-1 RNA duplex was obtained by integrating NMR, cryo-EM, and Molecular Dynamics
Figure 3. Atomic model of HIV-1 RNA duplex was obtained by integrating NMR, cryo-EM, and Molecular Dynamics
Creative Biostructure offers the cutting-edge instruments at our analytical center, a combination of high-resolution electron microscopy, and X-ray Crystallography and NMR Platform brings exceptional confidence to even the most challenging project. More importantly, an experienced team of professionals works closely with the client in experimental design, data analysis and 3D model reconstruction, delivering quality service for each and every project. For more information, please contact us.
Ordering Process
References
- Hong-Wei Wang, Jia-Wei Wang. How cryo-electron microscopy and X-ray crystallography complement each other. Protein Sci. 2017; 26: 32-39.
- Yong Xiong, From electron microscopy to X-ray crystallography: molecular-replacement case studies. Acta Crystallogr D Biol Crystallogr. 2008; 64: 76-82.
- KaimingZhang, Sarah C.Keane et al. Structure of the 30 kDa HIV-1 RNA Dimerization Signal by a Hybrid Cryo-EM, NMR, and Molecular Dynamics Approach. Structure. 2018; 26: 490-498.

