Crystallization with Mutant Library Approaches
High-throughput screening methodologies are already used in structural biology to define efficient protein crystallization and expression conditions. Meanwhile, screening approaches have been extended to the optimization of genetic constructs for improved soluble protein expression. With similarities to the directed evolution strategies used in protein engineering, a target gene encoding a poorly expressed protein is mutated by truncation, fragmentation or site-directed mutation.
In structural biology, both protein expression and crystallization processes contain large numbers of experimental variables that interact in complex ways and outcomes are often difficult or impossible to predict. In these situations, the desired result can be achieved by searching through a high diversity of possible solutions rather than by rational design. Screening for optimal protein-expression conditions for a specific open reading frame (ORF) is a well-established approach. In contrast, the design of constructs for expression testing is usually performed in a rational manner based upon careful analysis of the primary sequence of the protein. Generation of combinatorial libraries of randomly mutated genetic constructs coupled to a phenotypic screen or selection, has long been used in protein engineering for improving characteristics such as thermostability, enzyme specificity and binding-protein affinity.
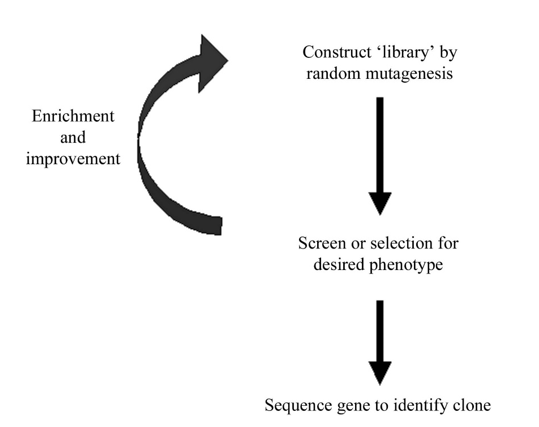 Figure 1. Schematic of the directed-evolution approach. A combinatorial library of randomly mutated genes is synthesized. Improved variants are identified via a high-throughput screen or selective process. Further improvements may be gained from iterative cycles of mutation and selection. Finally, clones are characterized by DNA sequencing to identify beneficial mutations.
Figure 1. Schematic of the directed-evolution approach. A combinatorial library of randomly mutated genes is synthesized. Improved variants are identified via a high-throughput screen or selective process. Further improvements may be gained from iterative cycles of mutation and selection. Finally, clones are characterized by DNA sequencing to identify beneficial mutations.
Using combinatorial library strategies, Creative Biostructure can improve protein solubility and develop an advanced and comprehensive tool for the structural biologist and these offer promising solutions to the production of difficult targets. These methods are well established in other fields, such as the improvement of industrial enzymes, and should transfer readily to problems in protein expression for structural biology. A mutagenic method is first used to generate diversity in the target gene and then a screen or selective process identifies the rare variants with improved protein-expression characteristics. Random mutation of the full-length gene and generation of DNA fragments are both strategies that can provide solutions to protein insolubility. An effective screen or selection process for identification of soluble constructs is required and this can either exploit an assayable activity of the target, as with an enzymatic activity screen, or may use a more generic readout such as a reporter fusion. The aim of the screen or selection is to rapidly reduce the large numbers of clones from a random library to a level where constructs can be assayed directly for solubility, usually by hand, using lysate fractionation and analysis by SDS–PAGE.
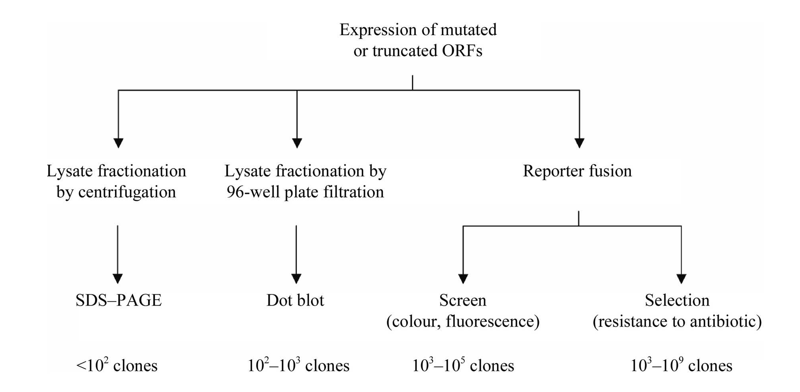 Figure 2. Overview of different methods of assaying for protein solubility.
Figure 2. Overview of different methods of assaying for protein solubility.
Creative Biostructure is a private R&D company specializing in protein engineering stability and structural biology. Provided protein optimization and crystallization service is unique thanks to integration of theoretical and experimental approaches from the fields of bioinformatics, molecular modelling, molecular biology, biochemistry, microbiology and biophysical chemistry. We employ proprietary state-of-art software tools and up-to-date automatic laboratory equipment to get biotechnologically interesting variants of proteins of interest while minimizing experimental effort, time and costs.
We like to be in touch with our customers so please feel free to contact us for a formal quote.
Ordering Process
Reference
- Darren J. Harta and Franck Tarendeaua. Combinatorial library approaches for improving soluble protein expression in Escherichia coli. Acta Crystallographica Section D. Volume 62| Part 1| January 2006| Pages 19-26. doi:10.1107/S0907444905036097.
