Protein Domain Prediction
Bioinformatics has become an important technique applied in many aspects of biology, such as genetics, genomics, system biology, and structural biology. It plays a pivotal role in the analysis of gene and protein expression and regulation. One goal bioinformatics pursue is protein structure prediction, which is highly important in biotechnology and drug design. In structural biology, bioinformatics finds wide applications in the simulation and modeling of DNA, RNA, proteins as well as biomolecular interactions.
 (National Marrow Donor Program)
(National Marrow Donor Program)
Protein sequence alignment
Comparison of protein amino acid sequences within a species or between different species can reveal
similarities between protein function, and relations between species. Multiple sequence alignments tools, such as Clustal W, Clustal X, T-Coffee, MAFFT, are
widely used. These tools often aid in identifying conserved region that can be used in conjunction with structural and mechanistic information.
Protein structure prediction
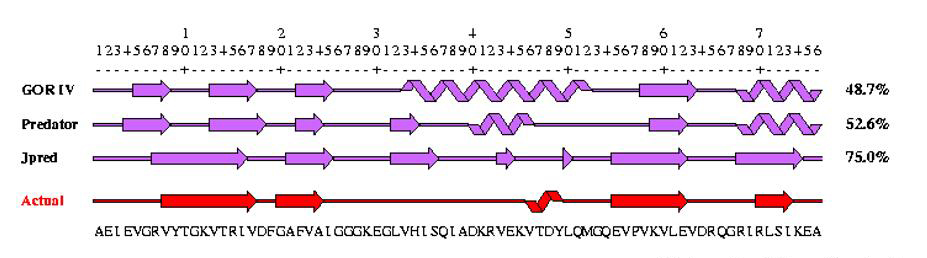
Protein structure prediction is another set of techniques in bioinformatics that aim to predict the folding, local secondary and tertiary structure of proteins based merely on their amino acid sequences. The best modern methods of secondary structure prediction in proteins reach about 80% accuracy. PSIPRED and JPRED are two of the most popular programs based on neural networks for protein secondary structure prediction, another notable program is GOR method, which is more successful in predicting alpha helices than beta sheets. Other commonly used software tools in protein secondary structure prediction and transmembrane helix and signal peptide prediction includes: SPIDER2 (the most comprehensive and accurate prediction tool to date), s2D, Meta-PP, HMMTOP, SignalP, etc. We provide protein domain structure prediction services, including secondary structure prediction, tertiary structure determination, and binding site analysis of proteins using many bioinformatics tools.
 (University College London)
(University College London)
Creative Biostructure aims to incorporate computational approaches into our structural studies of proteins. Our senior bioinformatics scientists will assist clients in protein sequence analysis, protein structure prediction, and protein-ligand interaction simulation. Please inquire online to see what we can do for you!
Ordering Process
References:
- Attwood, T.K., Gisel, A., Eriksson, N.E., Bongcam-Rudloff, E. (2011) “Concepts, historical milestones and the central place of bioinformatics in modern biology: a European perspective”. Bioinformatics-Trends and methodologies, 1:3-38
- Pirovano, W., Heinga, J. (2010). “Protein secondary structure prediction”. Methods in Molecular Biology.609: 327-348
- Larkin, M.A., (2007). Clustal W and Clustal X version 2.0. Bioinformatics, 23(21): 2947-2948
