Characterization of Protein-Protein Interactions
The interaction between two or more proteins (PPI) is the core of most biological processes. These interactions have been proven to be very diverse in thermodynamics, dynamics, structure, and regulation. Nuclear magnetic resonance (NMR) is a powerful technique for studying protein-protein interaction in solution. NMR spectroscopy has many valuable uses as a tool for studying PPI.
As an expert in the field of nuclear magnetic resonance, Creative Biostructure provides characterization services of protein interactions for our customers. We can provide customers with structural, kinetic, and thermodynamic information from NMR measurements of protein-protein interactions. The characterization of these interactions not only contributes to our understanding of biology, but also increasingly helps to design molecules that regulate protein-protein interaction (PPI).
Why Use NMR Spectroscopy
Nuclear magnetic resonance can,
- Provide specific information about the physical properties of a single functional group, such as ionization state, pKa, and hydrogen bond.
- Provide specific position information of dynamic motion of skeleton and side chain in solution.
- It is used to determine the contact between a single atom of a protein and its binding partner, and to study the dynamics of ligand binding.
Our Methods
Chemical shift perturbations (CSPs) analysis
Nuclear Overhauser Effects (NOEs)
Residual Dipolar Couplings (RDCs)
Paramagnetic Restraint
Solid-state NMR
Low-abundance species analysis
Relying on the above technologies. We can help our customers to deepen their understanding of the mechanism of protein-protein interactions.
Binding Site Analysis Services of PPI
Due to the changes in the physical and chemical environment of the protein interface, the formation of a protein complex (or any other chemical exchange process) will cause chemical shift perturbations (CSP), so chemical shift mapping is a sensitive indicator of protein binding sites. We provide a variety of analysis services.
Analysis service based on 15N-HQSC
We can use 15N-HSQC technology to complete PPI detection experiments for customers. From the 15N-HSQC titration experiment, a large amount of information about binding affinity and kinetics can be collected, which will form the basis for further analysis of the complex. The features of 15N-HSQC technology include,
- High sensitivity and good signal dispersion.
- Simplicity and economy of 15N marking.
- Good sampling of the interaction surface.
Analysis of PPIs involving large protein complexes
For large protein complexes, we use site-specific 13C-labeling of metal-bearing and aromatic side chains to label protein interaction sites. For large single peptides and large multi-subunit complexes, we use the "divide-and-conquer" strategy to allocate smaller units through traditional methods and then transfer the allocation to larger combinations. Then use 1H/13C CSPs to draw the interaction diagram with the partner/substrate protein.
Analysis of complex PPIs
We can also reveal more complex protein-protein interactions through CSP analysis, such as
- There are two or more combining points in fast exchange.
- The collection of specific complex and low-affinity non-specific complex in exchange.
- A compound exchanged between two conformational states in a rapid exchange.
Structural and Kinetic Analysis Service of PPI
Nuclear Overhauser effects analysis service
One of the most valuable sources of structural information used to define the binding interface is the nuclear Overhauser effect. We can provide "Straight" NOE and Transferred NOEs (trNOE) analysis services.
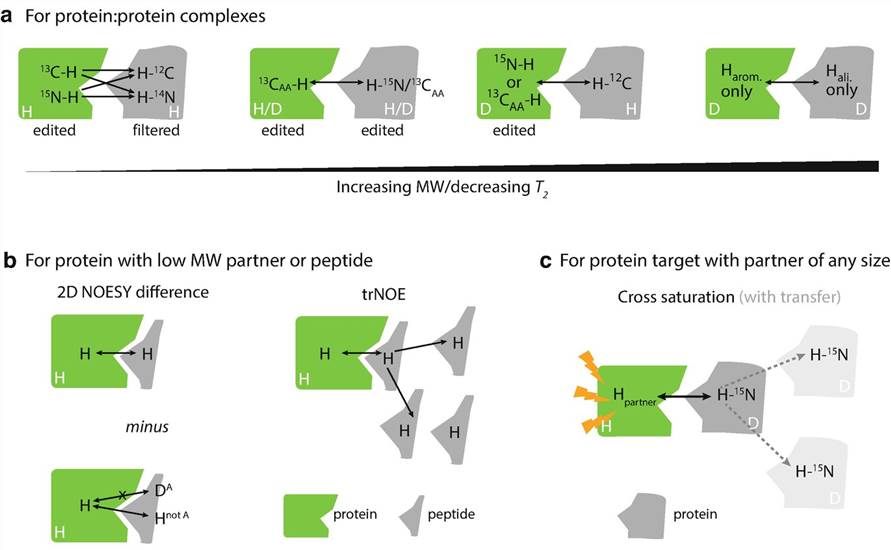 Figure 1. Isotope selecting and editing schemes for NOE /cross-saturation experiments (Gell et al., 2018)
Figure 1. Isotope selecting and editing schemes for NOE /cross-saturation experiments (Gell et al., 2018)
Residual dipolar couplings analysis service
For protein-protein complexes, RDCs can produce the relative orientation of a single subunit. This long-range information provides a valuable supplement to the local distance information provided by NOEs and CSPs.
Paramagnetic restraints analysis service
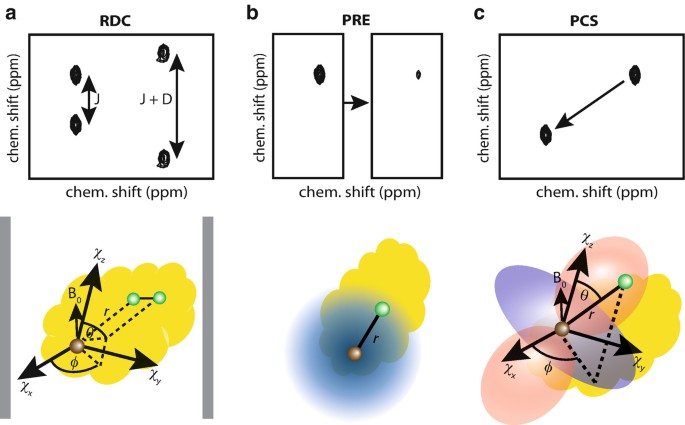 Figure 2. Schematic representation of RDC, PRE, and PCS experiments (Gell et al., 2018)
Figure 2. Schematic representation of RDC, PRE, and PCS experiments (Gell et al., 2018)
Provide various long-distance (up to 35 Å) and directional constraints. Here, we focus on the structural analysis of protein complexes using paramagnetic relaxation enhancement (PRE) and pseudo-contact displacement (PCS).
Creative Biostructure is committed to providing high-quality NMR analysis services to advance the life sciences fields. If you have any questions or needs, please contact us and our customer service staff will help you the first time.
Ordering Process
Reference
- Gell D A, Mackay J P. NMR spectroscopy in the analysis of protein-protein interactions. Modern Magnetic Resonance. 2006: 1339-1346.
